1. INTRODUCTION
Chronic hepatitis B virus infection (CHB) is a complex and dynamic disease related to the interaction between immune response and viral factors [1-3]. Based on the presence of HBeAg, HBV DNA levels, alanine aminotransferase (ALT) values, and the presence of liver inflammation, CHB is classified in different phases including immune tolerance (IT), HBeAg positive chronic hepatitis B (CHBe+), inactive carrier (IC), HBeAg negative chronic hepatitis B (CHBe-) [4].
Hepatitis B surface antigen (HBsAg) was discovered in 1968 and served as a diagnostic marker to identify HBV infection [5]. It is synthesized by three pathways including transcription from HBV genomic DNA, transcription from covalently closed circular HBV DNA (cccDNA), and integration from the S gene [6].
Recent evidences suggest that the level of serum quantitative HBsAg reflects the level of intrahepatic cccDNA and the transcription activity, as well as reflects the number of infected hepatocytes [7-10]. Many studies observed the serum HBsAg levels in different phases of CHB, had reported that serum HBsAg level is highest in immune tolerance phase (IT), slightly and continuously decreases in HBeAg positive immune clearance phase (CHBe+) and HBeAg negative reactivation phase (CHBe-) and lowest in inactive phase (IC) [11-15]. In addition, HBV DNA were different during various phases, the significant correlation with HBsAg has been reported in recent studies.
However, there was not a consistent correlation of serum HBsAg and HBV DNA in various phases with different genotypes [11, 13-15]. Positive correlations between HBsAg titers and serum HBV DNA, and cccDNA in hepatocyte have been noted in most studies of HBeAg positive patients [16]. However, studies in HBeAg negative CHB patients could not confirmed the correlation between the serum HBsAg and intrahepatic cccDNA [17, 18]. In CHB infected chimpanzees, Woodall C et al. had found that viral integration was modest during the HBeAg positive phases but had dramatic increase in HBeAg negative phases. This group had stated that only 10% of the mRNA in livers of HBeAg negative chimpanzees was derived from the HBV minichromosome but >90% was derived from integrated HBV sequences [19].
The disparity between HBsAg levels in HBeAg positive and HBeAg negative subpopulations in previous studies might also originate from the difference of HBsAg levels among HBV genotypes [17, 18]. Serum HBsAg were not different between genotype D and A in European CHB patients, but a clear difference of HBsAg levels was reported between genotype B and genotype C Asian CHB patients [11]. The quantitative measurement of HBsAg was developed in the recent decades and had quickly become an important tool to predict disease activity, to differentiate of immune tolerance and immune clearance in HBeAg positive patients and to predict inactive disease and spontaneous HBsAg seroclearance in HBeAg negative patients.
Numerous studies have addressed the use of HBsAg serum levels to understand the natural history of HBV infection and monitor the treatment response. On PEGylated interferon alpha (pegIFN) therapy, quantitative HBsAg could be applied to identify patients with a high probability of treatment response and to decide discontinuing in non-responders [20-24]. HBsAg level in serum also decline on nucleo(s)tide analogues treatment after two years and decline significantly in patients treated with tenofovir [25].
Our study aimed to describe the serum quantitative HBsAg and to evaluate the correlation between quantitative HBsAg and serum HBV DNA among Vietnamese CHB patients of different CHB phases.
2. MATERIALS AND METHOD
In this cross-sectional study, we recruited 276 aldult CHB patients who had not been taking antiviral therapy and had been visiting the University Medical Center (UMC) of University of Medicine and Pharmacy (UMP) at Ho Chi Minh City, Viet Nam from June 2013 to June 2016. Patients with hepatitis C virus (HCV) coinfection, alcohol hepatitis, cirrhosis, hepatocellular carcinoma, autoimmune disorders or under immunosuppressive treatment were excluded. Patients’ history, serial liver enzymes, HBeAg status, and HBV DNA were also recorded for categorizing the patients into five groups of HBV infection including Immune tolerance (IT), HBeAg+ Chronic Hepatitis (CHBe+), Inactive carrier (IC), Viral reactivation (VR), and HBeAg-Chronic Hepatitis (CHBe-) (Table 1). The upper limit of normal (ULN) of serum ALT was defined as 40 IU/L. We had added the VR group with HBV DNA > 104 cps/ml and persistence normal ALT to observe the difference from the VR and the CHBe-group.
Serum samples for analyses of quantitative HBsAg and HBV DNA were collected before antiviral treatment if treatment was indicated.
Serum HBV DNA was extracted by BOOM method and was measured by the in house Real-time PCR with the detection limit of 300 copies/ml.
HBV genotyping was performed using the Nested PCR to identify six genotypes from A to F. The molecular tests were all done at the Biomolecular Medicine Center of UMP at HCMC.
HBsAg quantification was analyzed by ECLIA (Electro-Chemi-Luminescent Immuno-Assay) using Elecsys HBsAgII Quant reagent (Roche) with automatic onboard dilution and the range of measurement from 0.05 to 52,000 IU/ml at the UMC at Ho Chi Minh City.
Data was analyzed using SPSS 16.0 software. Continuous variables with abnormally distributed values were expressed as median and interquartile ranges (IQR) and were compared by Mann-Whitney U or Kruskal Wallis test (≥3 groups). Correlation between serum HBsAg and HBV DNA values was analyzed by Spearman correlation coefficient. The significant difference was set at p value <0.05.
3. RESULTS AND DISCUSSION
A total of 276 patients were recruited. These treatment-naïve patients were categorized in 5 groups including immune tolerance (IT) (n=64), HBeAg positive chronic hepatitis (CHBe+) (n=66), inactive carrier (IC) (n=56), viral reactivation (VR) (n=47) and HBeAg negative chronic hepatitis B (CHBe-) (n=43). The baseline characteristics of the study population was presented in Table 2.
There were more male (63.7%) than female patients and more patients infected HBV genotype B (69.3% with genotype B and 5.3% with mix genotype B and C) than genotype C in this study population. A male predominance was observed in active hepatitis or active replication groups: CHBe+ (72.7%), CHBe-(76.7%) and VR (70.2%). HBeAg negative groups had more rate of older than 40 age groups compared to HBeAg positive groups (p<0.001). There was significant higher rate of genotype C (including mix genotype C and B) in CHBe+ group than in other groups (42% compared to 20 to 25%, p=0.033).
The median value of serum HBsAg was not significantly different among subgroups of CHB patients with genotype B and genotype C (data not shown).
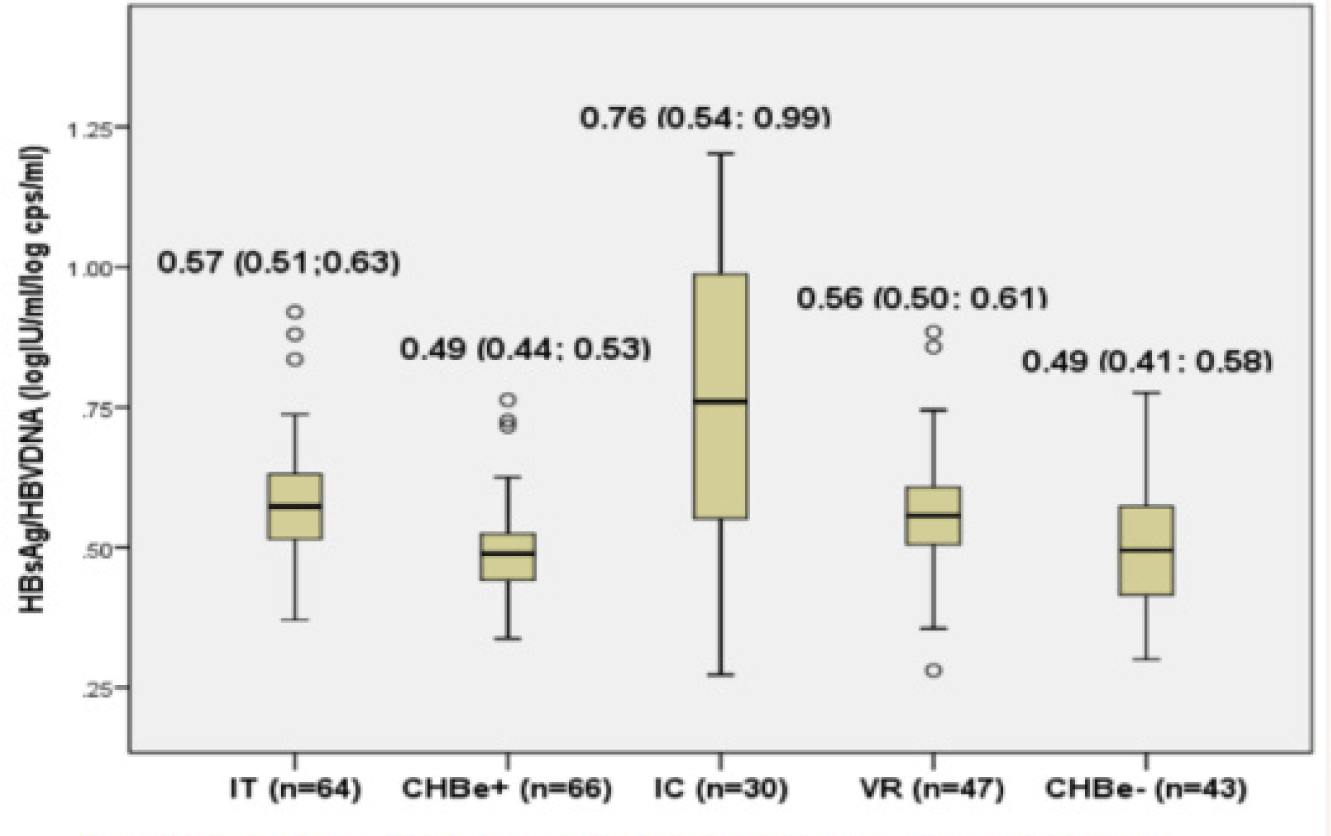
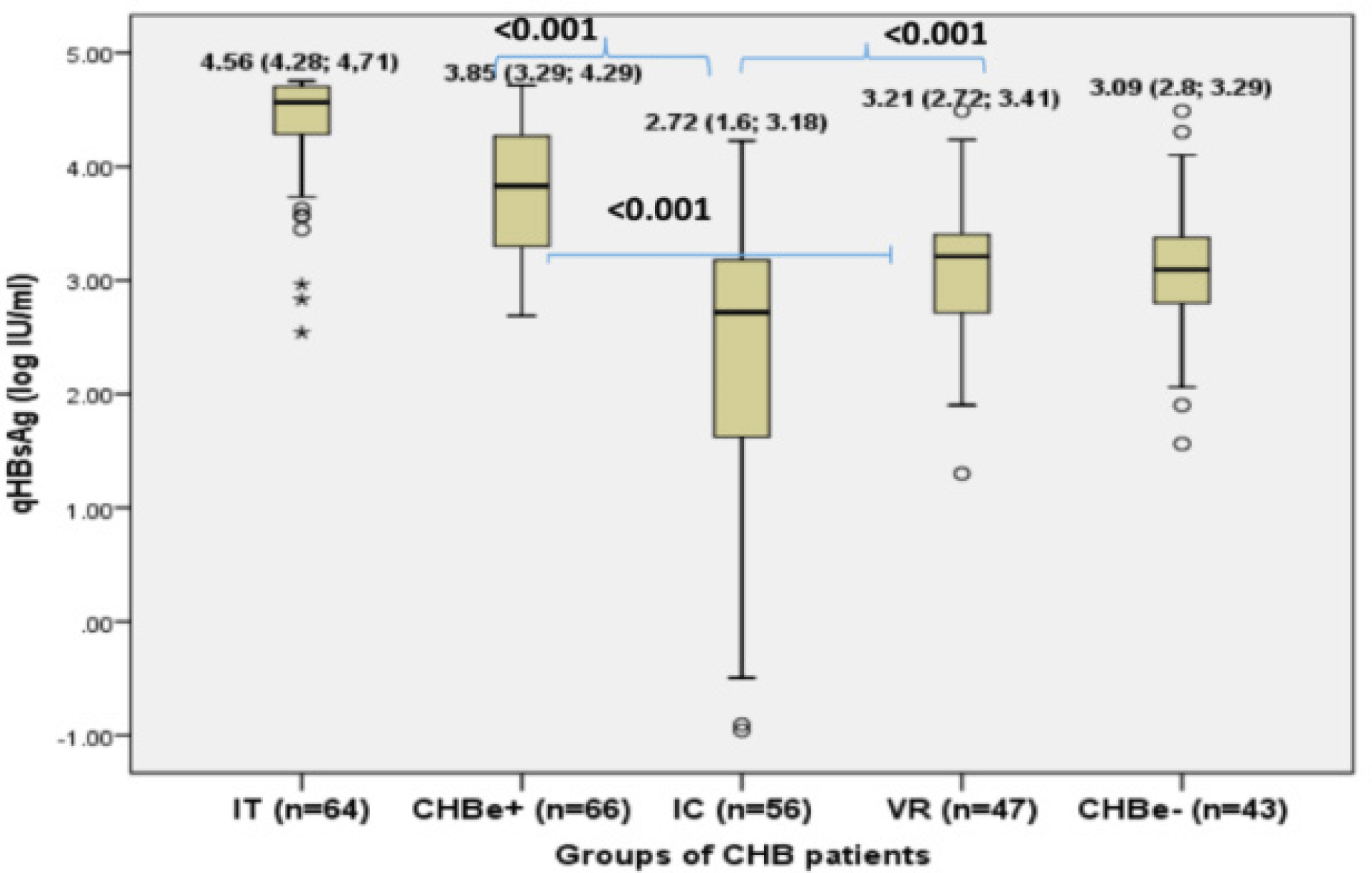
The distribution of HBsAg in different study groups was presented in Figure 1. The median serum HBsAg was highest in IT group (4.56 log10 IU/mL, mostly over 4 log IU/ml) and in CHBe+ (3.85 log10 IU/mL and mostly 3-4 log IU/ml). The serum HBsAg in HBeAg positive groups were significantly higher than in HBeAg negative groups (most of under 3.5 log IU/ml): IC (2.72 log10 IU/mL), VR (3.21 log10 IU/mL), CHBe-groups (3.09 log10 IU/mL). The IC group had the lowest level of serum HBsAg and especially had a wide distribution (Figure 1).
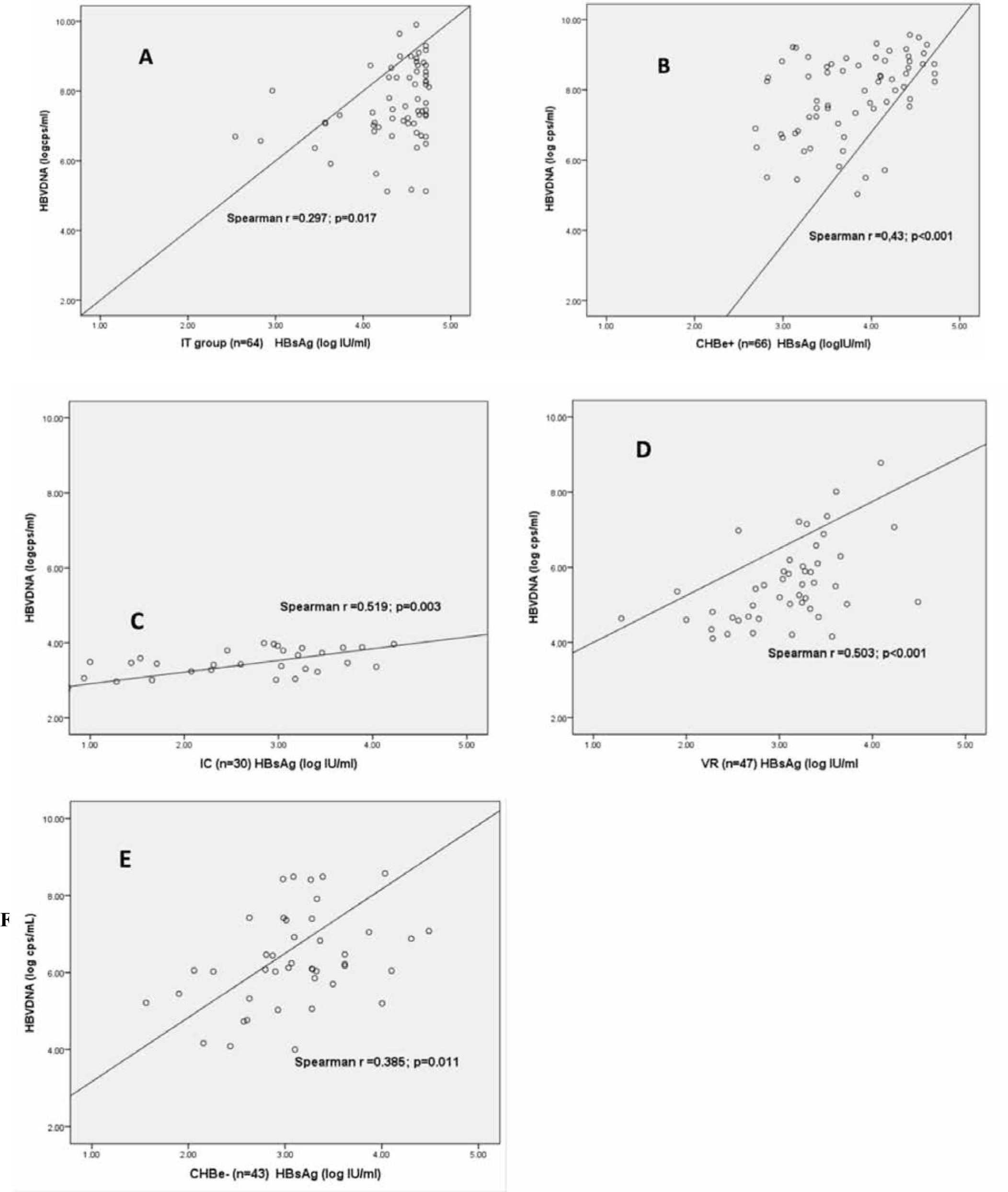
There was positive correlation between HBsAg and HBV DNA in overall study population (Spearman r=0.615, p<0.001), in HBeAg positive (r=0.26, p=0.003), HBeAg negative groups (r= 0.4, p<0.001) and in all five groups of CHB patients in this study: in IT group (Spearman r = 0.297, p=0.017), in CHBe+ (r=0.43, p < 0.001), in IC group (r=0.519, p=0.003), in VR group (r=0.503, p<0.001) and in CHBe-group (r=0.385, p=0.011). The correlation was high in CHBe+, IC, VR group and moderate in CHBe- and IT group.
The HBsAg/HBV DNA ratio were around 0.5 in all groups of patients except in the IC group. There was a significant higher HBsAg/HBV DNA ratio in IC group compared to IT, CHBe+, VR and CHBe-groups (0.76 vs 0.57, 0.49, 0.56, 0.49) (Figure 2). The ratio of HBsAg/HBV DNA was widely distributed in the IC group (data not shown).
This study evaluated the baseline serum quantitative HBsAg and HBsAg/HBV DNA ratio in different stages of HBV infection. Most of patients had infected with genotype B and C. We found that serum HBsAg levels and HBsAg/HBV DNA ratio were different throughout various stages of chronic HBV infection but not different between genotype B and C. Data from our study also showed that IT phase has the highest median value of HBsAg, while IC group has the highest HBsAg/HBV DNA ratio. Moreover, we demonstrated a significant correlation between HBsAg and HBV DNA among the five groups of CHB.
In our study, the median value of HBsAg was highest in the IT; lower in other late evolution groups. These observation had also been stated in other studies [11-13, 15, 26]. The median HBsAg of all CHB groups in our study is consistent with other studies in Asian populations [11, 14, 15, 26]. However the median of HBsAg in our IT and CHBe+ groups was lower than in European population [13] due to our lower selection criteria for HBV DNA (> 5 log cps/ml) and lower peak HBV DNA value of these groups.
It was mentioned that cccDNA transcription and viral replication were highly active accompanied with highest serum HBV DNA in the early immune tolerant (IT) and IC (CHBe+) phases. In these HBeAg positive phases, HBsAg from cccDNA (HBsAg ER-secretory pathway) and from virion (DNA replication pathway) origin were also highly secreted [27].
In the HBeAg negative phases, HBsAg secretion from these 2 above pathways were reduced due to the immune activation aim to control HBV replication after HBeAg seroconversion. However the synthesized of HBsAg that originated from double stranded linear (DSL) DNA form (integrated DNA pathway) become more often [19]. Therefore, HBsAg from this source were inconsistent with HBV replication and its high levels was not well correlated with serum HBV DNA levels in these late HBeAg negative phases.
In our data, one half of patients in our IC group had lowest HBsAg values that reflected lowest HBV replication without or none of HBsAg from integrated DNA source. This is in agreement with other studies that HBsAg <3log IU/ml with or without of HBV DNA <4 log IU/ml had been considered inactive carriers states in other studies [11, 13, 28].
Chan HLY. et al had also stated in IC patients the slow rate (0.043 log IU/mL/year) but steadily clearance of HBsAg reflected the stablility of immune control [12].
The level of HBsAg was lowest in our IC groups. This result was consistent with other studies in HBeAg negative patients, but was higher than that from Kim’s study, in which more cases with older age and cirrhosis were included [15]. Kim YJ. and his group also proved that age had a significant negative correlation with the HBsAg level.
The significant higher HBsAg level in our VR group and CHB HBeAg negative groups in comparison to IC group reveal that viral reactivation and HBsAg secretion were paralelly increased in these HBeAg negative group.
The Spearman r co-efficient correlation between HBsAg and HBV DNA among all groups of study were varied between 0.3 and 0.5 and were significant in all groups. In this study the lower correlation was seen in IT and CHBe-group, higher in CHBe+, IC and VR group. The different correlation in different phases of CHB had also been observed in other studies. Jaroszewicz found moderate correlation in all phases of genotype D patients [13]. Kim YJ. found strong correlation in all phases of genotype C patients except CHBe negative group [15]. Antaki N. reported strong correlation in IT group, moderate in CHBe-group, and no correlation in CHBe+ and IC in genotype D population [29]. Zeng L-Y. showed strong correlation in CHBe+ phase, moderate in CHBe- and IT, poor in IC in genotype B or C population [26]. Karra VK. found strong correlation in the IT and CHBe+ phase, moderate in IC and weak correlation in CHBe- [14].
In our subgroup of IC, the correlation was moderating but we had observed the highest ratio of HBsAg/HBV DNA. We also found that the values of HBsAg in this IC group was widely distributed in spite of the low variation on HBV DNA values. We had discovered that we could divide patients in IC to 2 separate subgroups: the first half of patients with low HBsAg (<3 log IU/ml) and low HBV DNA values (<3 log copies/ml) which had normal ratio of 0.5 (left part of fig 2C). The second half with high HBsAg values (>3 log IU/ml) and low HBV DNA levels (<3 log copies/ml) (right part of fig 2C). Molecular studies in chimpanzee had also state on the HBsAg production from the integrated HBsAg sequences in this stages of CHB infections [19]. We suggested that the high HBsAg/HBV DNA ratio in this IC groups might present for (1) the patients who had recently had HBeAg seroconversions with HBsAg was decreasing; (2) the early viral reactivation patients; and (3) the group with non-cccDNA or integrated DNA HBsAg production.
In the situation (1), the production of Dane particle had stopped, the HBsAg in the outer layer were likely to decrease accordingly. In case of excess HBsAg was still produced from cccDNA (HBsAg pathway) or from integrated DNA pathway, the HBsAg/HBV DNA ratio increased due to relatively high HBsAg.
The technique of quantification HBsAg employed in this study could measure but could not distinguish 3 kinds (S, M, L) of surface protein. Moreover, the low HBV DNA criteria to classify the IC group automatically excluded a number of HBV DNA negative patients in the analysis of correlation between HBsAg and HBV DNA. Serum HBsAg and correlation between and HBV DNA in the HBeAg negative groups need to be study in the aspect of HBV DNA integration.
A cohort study will recognize the well controled replication (low or negative HBV DNA) patients that not had enough time for significantly reduced of HBsAg and patients with high and lonely HBsAg secretion from the integrated DNA but not reactivated replication that had low or undetected HBV DNA.
The limitations of our study were the cross-sectional design and the low number of HBV DNA positive patients in the IC group. In addition, the low HBV DNA viral load (105 cps/ml) in IT group may be a bias factor to clarify the immune tolerance or immune clearance status in the HBeAg positive group. Further and larger sample size studies are needed to evaluate the value of HBsAg/HBV DNA ratio in HBV DNA estimates for the lower cost if applicable.
4. CONCLUSION
Our study demonstrated that serum HBsAg level fluctuated during the natural phase of CHB infection but was not significant different between genotype B and C. There were significant correlations between HBsAg and HBV DNA in all five phases of CHB suggesting a role of HBsAg in CHB classification. The wide distribution of HBsAg in the IC group raised the question on the existence of HBsAg integration in CHB patients.