1. INTRODUCTION
In addition to conventional cancer treatments such as surgery, radiotherapy, and chemotherapy, the trend toward combining conventional treatment and natural-derived anti-cancer extracts is promising [1, 2]. The combination of natural products with conventional treatment to overcome the current cancer resistance is on the rise [3]. Currently, more than half of humanity does not have access to modern medicine and relies on traditional treatments [4]. A recent analysis of the strategies used in the discovery of new medicines showed that 36% of the first-in-class small-molecules approved by U.S. Food and Drug Administration (FDA) between 1999 and 2008 were natural products or natural products derivatives [5]. Recent studies have shown that C. aromatica is effective in reducing tumor growth, glioma in brain cancer, and inhibiting the development of liver tumors in animal models [6, 7]. C. aromatica extract also has effects on colorectal cancer [8], as well as preventing progression to chronic oesophagitis or esophagus cancers such as Barrett’s disease [9].
The species of Curcuma are usually found in tropical and subtropical forests, margins of the forest, open grasslands, secondary forests, plantations, areca nut groves in many countries such as India, China, Thailand, Malaysia, Philippines, Vietnam, and Indonesia [10]. In Vietnam, C. aromatica, “Ngải trắng” in Vietnamese, is found in the forests of the Northwest, Quang Binh, Daklak, An Giang, and so on, but only the essential oil from rhizomes collected in Northern Vietnam has been analyzed in previous studies [11, 12]. The rhizome morphology of Curcuma plants is very similar, making it difficult to perform correct identification of each type. Minami et al. conducted a study to identify species of Curcuma species collected from Japan, Vietnam, Taiwan, Indonesia, Thailand, and China, in which the three most distinguishable species were identified as C. aromatica, C. longa, and C. zedoaria.
Up to now, there are some studies that have reported the composition of C. aromatica containing about 6% of essential oils, including: 1,8-cineol, p-cymene, curcumin, β-curcumene, curdione, neocurdione, curcumol, tetramethylpyrazin, 1,2- hexadecanediol, 9-oxo-neoprocurcumenol, neoprocurcumenol, xanthorrhizol, germacrone, camphor, curzerenon, 7-methanoazulen, β-elemene, and linalool [13, 14]. In particular, curdione and germacrone are compounds which have been shown to have anti-tumor effect, anti-inflammatory, antiviral, and antioxidant activities. Curcumin is well-known in providing good therapeutic benefits on a variety of human diseases such as inhibiting the cell growth of various cancer cell lines, inducing apoptosis as well as effecting the cell-cycle regulation of cancer cells [15-17]. It has been determined in the previous study that there is a difference in the curcumin content among individuals of the Curcuma species [18]. Curdione and germacrone are the well-known anti-inflammatory and anticancer agents [18-21]. Therefore, the objectives of the present study were to identify the collected samples according to their trnSfM sequences and compare the curcumin, curdione, and germacrone contents from the samples collected in An Giang province, Vietnam.
2. MATERIALS AND METHOD
Six rhizome samples supposed “Ngải trắng” named NT1, NT2, NT3, NT4, NT5, and NT6 were collected in An Giang province, Vietnam by two traditional medicine experts. All samples were deposited in the Center for Molecular Biomedicine, University of Medicine and Pharmacy at Ho Chi Minh City, Vietnam and planted for seed sampling. Curdione and germacrone were the working standards (purity > 98%) [22]. Curcumin was purchased from Institute of Drug Quality Control Ho Chi Minh city (purity > 92%).
Morphological characteristics of six samples including the shape, color, size, and texture were observed and examined.
Fresh rhizomes were cut into thin slices, stained in a combination of carmine alum and iodine green. The anatomical characteristics were photographed, described and analyzed.
Total DNA of 6 samples was isolated from 50-100 mg of fresh tissue of rhizomes using DNeasy Plant Mini Kit (QIAGEN, Germany). The amplification of the trnS-trnfM region of cpDNA from all specimens was carried out using primer trnSfM (Table 1). The PCR was performed in the reaction mixtures of 50 μl. The mixture contained TaKaRa LA Taq DNA Polymerase (Takara, Japan), 10 pmol primer, and 20-50 ng total DNA. Amplification was performed in a DNA thermal cycler (EppendorfMastercycler Nexus, Eppendorf, Germany). The PCR conditions used were as follows: 98°C for 2 minutes followed by 35 cycles of 15seconds at 98°C, 30 seconds at 55°C, 90 seconds at 68°C, and a final extension for 7 minutes at 72°C. Products of PCR were separated electrophoretically on a 2% (w/v) agarose gel in 1X TBE buffer and photographed under ultraviolet (UV) light.
| Locus | Primer | Sequences (5’– 3’) | Length (bp) | Annealling temp (°C) | Reference |
|---|---|---|---|---|---|
| trnS-trnfM | trnSfM-f | GAGAGAGAGGGATTCGAACC | 1475 | 62 | Minami et al 2009 |
| trnSfM-r | CATAACCTTGAGGTCACGGG |
The PCR products were purified using ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific) and subsequently sequenced with forward primer by BigDye® Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). All resultant sequences of the trnS-trnfM region were aligned by using Genescan. The nucleotide sequence of the trnS-trnfM region (trnSfM) was detectable since it was a suitable candidate for the molecular identification of C. longa, C. aromatica, C. zedoaria, and C. xanthorrhiza [18].
Curcumin qualitative analysis was carried out using a published method [23] with slightly modification. An HPLC system (Azura, Knauer, Germany) was empolyed. The separation was performed on a Syncronis C18 column (250 x 4.6 mm; 5 μm) (Thermo Scientific, USA) Mobile phase was a mixture of acetonitrile and phosphoric acid 0.07% in which the ratio of acetonitrile was at 50%. The flow rate was 1 ml/min. Detection was performed at a wavelength of 428 nm at 30°C. The sample injection volume was 20 μl.
Curcumin reference solution: curcumin (1.0 mg) was dissolved and diluted in a 10 mL volumetric flask by methanol.
Sample solutions: rhizome powder sample (0.5 g) was extracted by 15 ml of methanol using heat under reflux method for 30 min (repeated 2 times) and filtered. The filtrate was evaporated, and the residue was then dissolved and diluted in a 10 mL volumetric flask by methanol.
Curdione and germacron qualitative analysis were carried out as previously described [24] with slightly modification using by an Azura HPLC system (Knauer, Germany). The previous method was modifewas modified. The separation was performed on a Syncronis C18 column (250 x 4.6 mm; 5 μm) (Thermo Scientific, USA). Mobile phase was mixtures of acetonitrile and water with a gradient program. The ratios of acetonitrile were 10 %, 20 %, 48 % and 90 % at 0 min, 10 min, 52 min and 90 min, respectively. The flow rate was set at 1 ml/min. Detection wavelength was set at 214 nm. The column temperature and the sample injection volume were set at 30°C and 20 μl, respectively.
Germacrone reference solution: germacrone (10 mg) was dissolved and diluted in a 10 mL volumetric flask by methanol.
Curdione reference solution: curdione (10 mg) was dissolved and diluted in a 10 mL volumetric flask by methanol.
Sample solutions: rhizome powder sample (0.5 g) was extracted by 15 ml of methanol using heat under reflux method for 30 min (repeated 2 times) and filtered. The filtrate was evaporated, and the residue was then dissolved and diluted in a 10 mL volumetric flask by methanol.
3. RESULTS
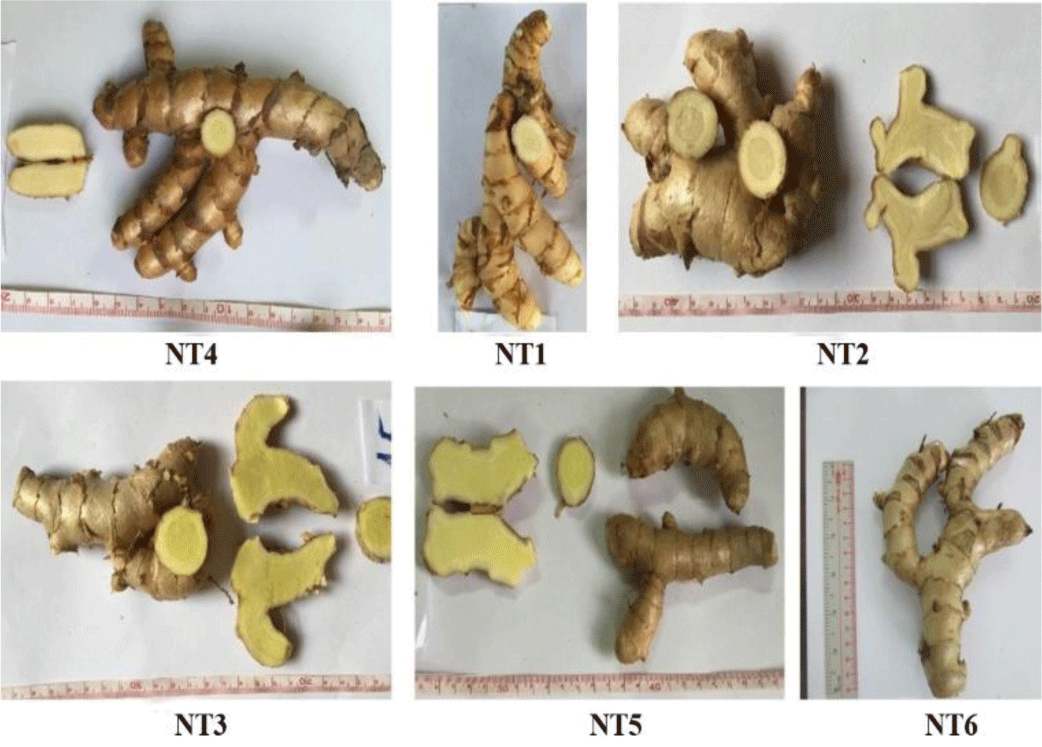
Figure 1 illustrates the rhizomes of six samples collected from An Giang province, Vietnam.
The rhizome of sample NT1 had a light brown color, cylindrical, branched in Y-shape. The main rhizome was 5 – 7 cm long and 3-5 cm in diameter. The branched rhizome was 6 – 15 cm long and 1 – 3 cm in diameter. In the outer layer, there were many scars of leaves. It was clearly observed that the cross-section included two parts: cortex and stele with yellowish-white and fibrous.
Morphological characteristics of samples NT2, NT3, NT4, NT5, and NT6 were very similar to those of sample NT1. The six collected samples were almost indistinguishable due to very little difference in the external characteristics and the taste of the rhizome.
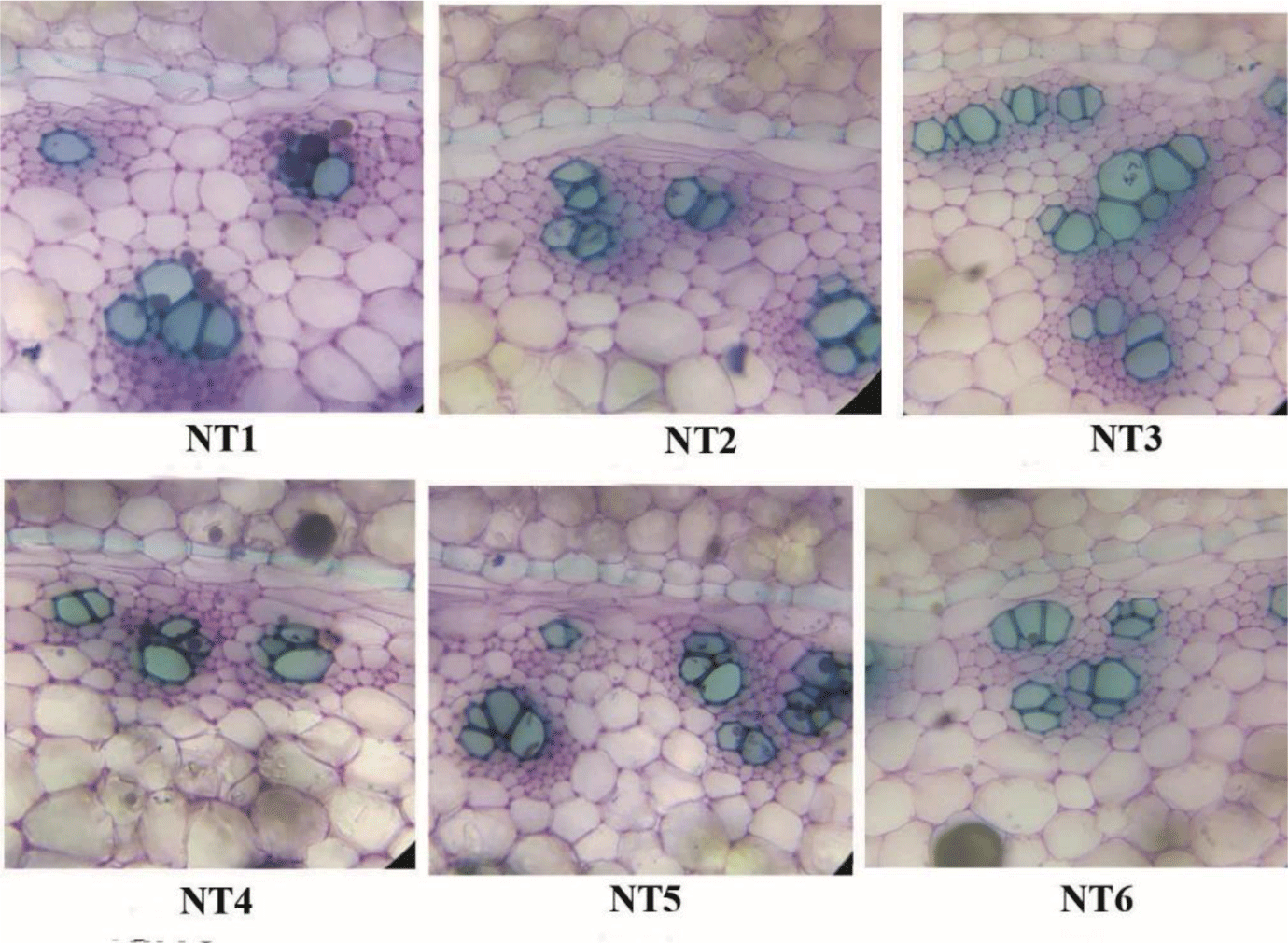
Anatomical characteristics of the transections of six samples were described and analyzed (Figure 2 and 3). The cross-section of sample NT4 (Figure 2) clearly consisted of two parts: cortex and stele. The cortex was broad. The epidermal cells were flat and had thin walls. There were 4-6 layers of flattened cork cells. In the parenchyma, leaf-trace bundles and secretory cells were scattered. Endodermal cells with distinct Casparian dots were obvious. Pericycle consisted of 1-2 rows of the parenchymatous cell. The stele had scattered vascular bundles, some more near the pericycle and lessened inward. Each vascular bundle had phloem outside the xylem. Parenchymatous cells packed with secretory cells and starch granules.

The anatomical characteristics of the other samples (NT1, NT2, NT3, NT5, and NT6) were found to be similar with these of NT4. The number of xylem vessels of the six samples was distinct. However, this difference could not distinguish the six samples. On the other hand, until now, there has been no report on the anatomical characteristics of C. aromatica. Therefore, it is difficult to distinguish and determine which sample was C. aromatica by using the anatomical data.
All the PCR products were approximately 1475 bp long and the sequencing results of trnSfM intergenic spacer region were shown in supplementary Figure 1. The partial DNA sequences in the trnSfM spacer of Curcuma species (with a length of approximately 230 bp) showed that there were two base substitutions, two base deletions, and different numbers of adenine - thymine (AT) repeats. In addition, the DNA polymorphisms at three sites (nucleotide positions 176, 207, and 216-232 from the 5’ end of the forward primer) divided the collected Curcuma species into three different haplotypes: G-(AT)8, A-(AT)6, and G-(AT)5. These samples were distinguished on the basis of the polymorphism at nucleotide 176 and the number of AT repeat in the 216–232 segments. The number of AT repeats in the C. aromatica had an 8-AT repeat (G-(AT)8), whereas the C. longa showed a 6-AT repeat (A-(AT)6 and those in C. zedoaria had a 5-AT repeats (A-(AT)5) (Table 2). Therefore, among total six samples harvested in An Giang province, Vietnam NT1, NT2, and NT3 were identified as Curcuma aromatica; NT4 and NT5 were identified as Curcuma longa while NT6 was identified as Curcuma zedoaria.
| Sample # | Halotypea | Nucleotide positionb | Size (bp) | Spieces | ||
|---|---|---|---|---|---|---|
| 176 | 207a | 216-232 | ||||
| NT1 | G-(AT)8 | G | _ | (AT)8 | 231 | C. aromatica |
| NT2 | ||||||
| NT3 | ||||||
| NT4 | A-(AT)6 | A | _ | (AT)6 | 227 | C. longa |
| NT5 | ||||||
| NT6 | G-(AT)5 | G | _ | (AT)5 | 225 | C. zedoaria |
Curcumin was identified as the peak with retention time at 15.8 min (Figure 4). There was no curcumin peak was detected in sample NT1, NT2, NT4 and NT6. However, NT3 and NT5 samples were found to contain curcumin in the HPLC chromatograms.
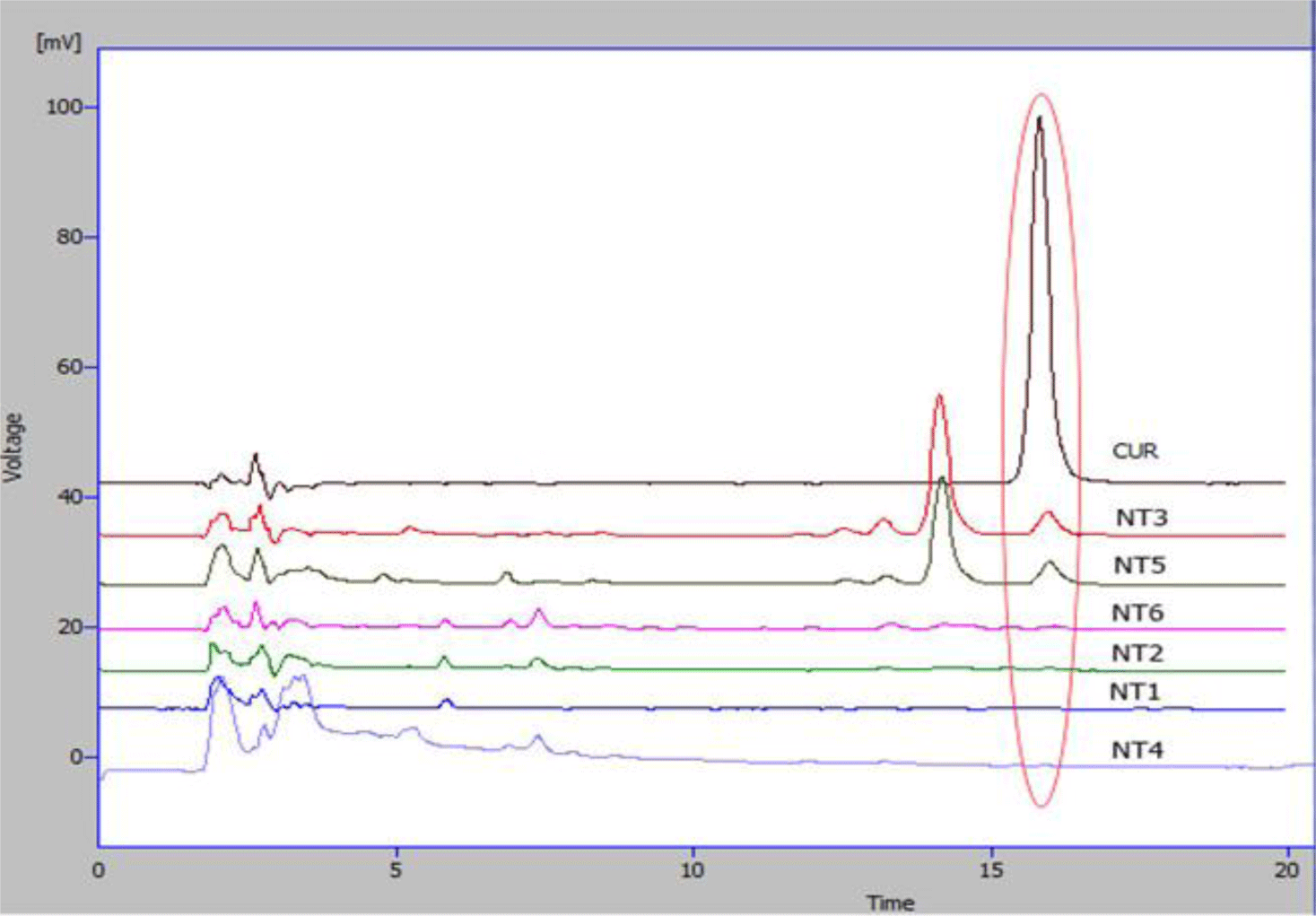
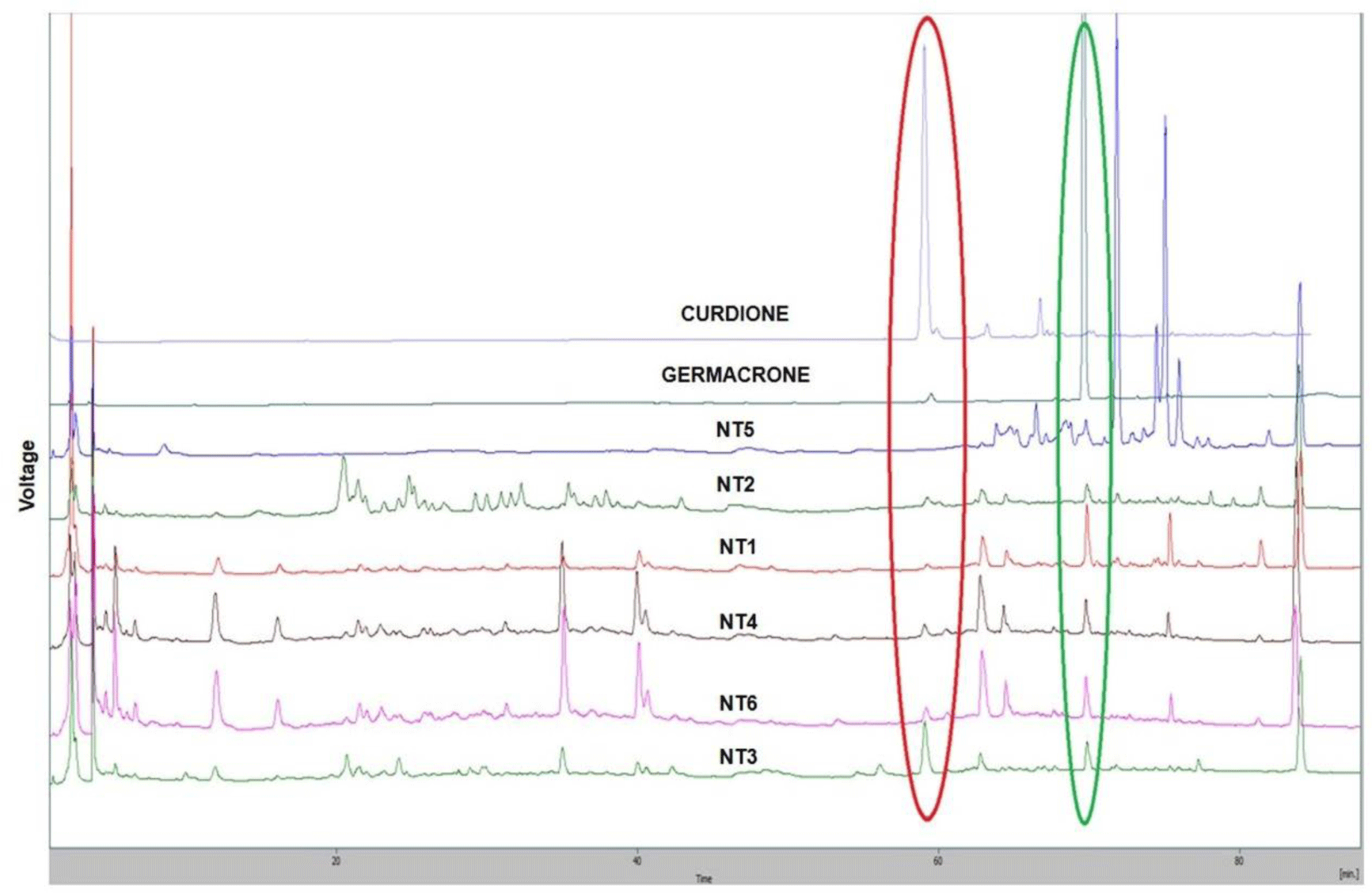
Figure 5 provides that all six samples had the germacrone peaks (Rt= 69.2 min) and curdione peaks (Rt = 58.5 min) except sample NT5 (the curdione peak could not be detected). However, there were differences in the ratios of germacrone and curdione among the six samples. Among the three samples identified as C. aromatica, NT2 was the sample with the lowest peaks of germacrone and curdione. NT1 showed the highest peak of germacrone but the lowest peak of curdione. NT3 was the sample with the highest peak of curdione and the high peak of germacrone.
4. DISCUSSION
C. aromatica is widely used in Vietnamese Traditional Medicine. According to our results, it is difficult to distinguish which species is C. aromatica only relying on the morphological and anatomical characteristics. In this study, the six Curcuma species examined are clearly identifiable by their trnSfM sequences. The AT repeats in trnSfM marker in Vietnamese C. aromatica samples were longer than in Japanese samples and this polymorphism contrast is due to geographical difference. A similar geographic difference was also found in C. zedoaria collected in Japan and China when Sasaki and colleagues used amplification-refractory mutation system analysis of the 18S rRNA gene [25]. Although the polymorphism at nucleotide 176 and the number of AT repeat in the 216–232 segments in the trnSfM region requires more complex steps, namely DNA sequencing, and time-consuming, this method can identify Curcuma species. The distinction of herbal medicine in the same family is very important for the identification and validation of the ingredients of the drug in the herbal medicine industry. Recently, Rafi et al. [17] used thin-layer chromatography (TLC) fingerprint analysis to identify Curcuma species by determining different regions with UV light (254 and 366 nm).
Curcumin has been well-known as the anti-inflammatory, antioxidant and anticancer compound [26]. Curdione and germacrone have been reported to possess significant anticancer activities [27]. The HPLC analysis results revealed that among the six samples named “Ngải trắng” in An Giang province, Vietnam two samples (NT3 and NT5) were found to contain curcumin, five samples (except NT5) contained curdione and all six samples contained germacrone. Therefore, the six samples named “Ngải trắng” in An Giang province, Vietnam could be the potential herbal medicine for anti-inflammatory or anticancer treatment.
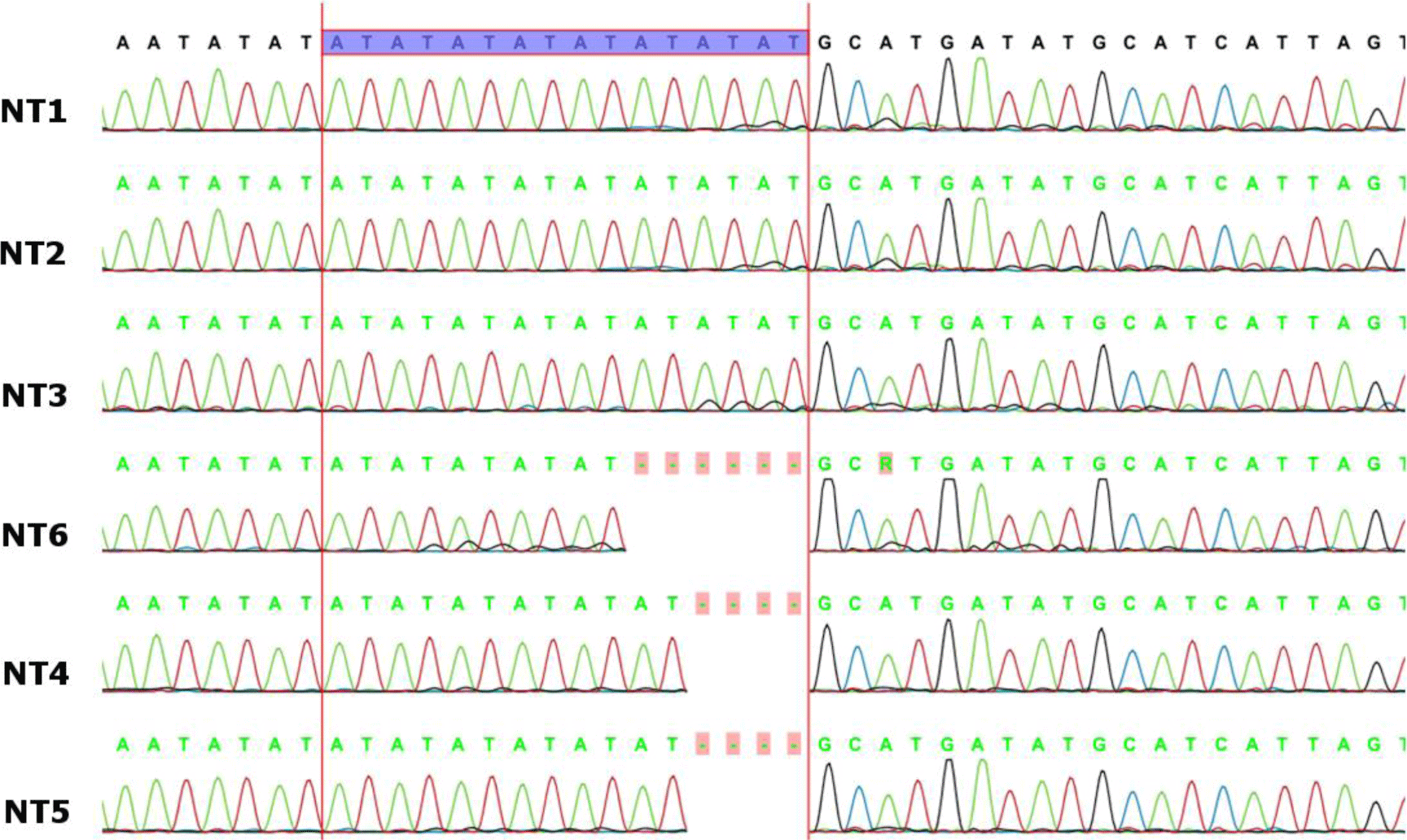
Interestingly, the three samples of C. aromatica (NT1, NT2, NT3) were the same in trnSfM the sequences; however, the contents of curcumin, curdione, and germacrone were different. NT2 was the C. aromatica sample contained the lowest contents of the three mentioned compounds. NT3 was the only C. aromatica sample that simultaneously contained curcumin, curdione and germacrone in its HPLC chromatographs. The differences in the ratios of the three active compounds could be explained by the variations of the natural conditions in the growing areas of the three samples identified as C. aromatica by Sanger sequencing.
5. CONCLUSION
DNA sequencing is a simple and accurate method for identifying C. aromatica with other similar species. The contents of curcumin, curdione, and germacrone were found to be different among the three C. aromatica samples. The NT3 C. aromatica sample growing in An Giang province, Vietnam which simultaneously contained curcumin, curdione, and germacrone could be the promising candidate for combination therapy in anti-inflammatory and cancer treatment.