1. INTRODUCTION
Pseudomonas aeruginosa is one of the most antibiotic-resistant bacteria which plays an important role in the inception of nosocomial infections all the word. These pathogens are common causative agents of pneumonia, bacteremia, urinary tract, skin and soft tissue infections [1]. The dissemination of antibiotic resistant genes between bacterial species in infectious diseases in a serious and worrying problem [2]. Carbapenem are type of β-lactam drug, which are often used as a last resort for the treatment of infections caused by drug resistant Gram-negative bacteria [3]. Carbapenem have brssoad spectrum of antibacterial activity and are used as last resort drugs for the treatment of infections caused by multiresistant P. aeruginosa isolates. Carbapenemase are classified according to a structural and a functional classification. Ambler's structural classification comprises three molecular classes: A) The extended-spectrum beta-lactamases (ESBL) that are inhibited by clavulanic acid, B) the metallo-beta-lactamases (MBL) and D) the oxacillinases (OXA) [4].
In Vietnam, according to a report at the Scientific Conference of author Doan Mai Phuong [5], the rate of resistance to imipenem (carbapenem group) of P. aeruginosa with the rate of three regions in the North: 12.9%, Central: 10.5%, South: 80% (n = 469) and the prevalence of carbapenem resistance genes was blaIMP: 46.2% (n = 145) and blaVIM: 0.7% (n=142). At Xuyen A hospital (2017) [6], the rate of P. aeruginosa resistant to ampicilline-sulbactam: 86.67%, amoxciline-clavulanic acid: 96.15%, cefoperazone-sulbactam: 20%, piperacillin-tazobactam: 23.21%, cefuroxime: 100%, ceftriaxone: 87.5%, ceftazidime: 20%, cefotaxime: 66.67%, ciprofloxacin: 36.84%, doxycyline: 74.07%, levofloxacin: 30.9%, meropenem: 29.62%, imipenem: 21.81%, colistin: 7.4%, amikacin: 16.07%.
Besides using carbapenem, the combination with colistin (polymyxin E) was selected as a last resort antibiotic in the treatment of multiresistant strains such as P. aeruginosa [7]. The continual increase in new strains of carbapenem resistant Pseudomonas aeruginosa (CRPA) requires more use of colistin, leading to the risk of colistin resistance is inevitable. In order to screen resistant phenotypes and identify the gene for carbapenemase production, research to implement inactivation method of carbapenemase (mCIM), colistin broth disk elution (CBDE) tests for colistin resistance and multiplex real-time PCR. Gene detected (i.e. Ambler classification) that can appear in epidemiological areas in Vietnam for P. aeruginosa are: class A: blaKPC, class B: blaIMP, blaVIM, blaNDM, and class D: blaOXA-48 [8].
Knowing the mechanism of resistance as well as classifying resistance genes helps clinicians choose appropriate antibiotics for effective treatment. It also helps to control infection, providing an optimal solution to prevent the spread of resistant genes. This study identified the prevalence of carbapenemase producing P. aeruginosa by the modified carbapenem inactivation method (mCIM) and distribution of common genes encoding carbapenemases of P. aeruginosa by multiplex real-time PCR technique. In addition, colistin broth disk elution test (CBDE) were utilized to detect the resistance of P. aeruginosa to colistin.
2. MATERIALS AND METHOD
All specimens (n=159) isolated from clinical samples (blood: 3,1%, pus: 18,2%, urine: 17%, sputum: 28,3%, body fluids: 33,3%) were involved in this study. They are identified and routinely used by an automated antibiotic machine (Phoenix ™ M50-Becton Dickinson-USA). Methods are developed based on the Institute of Clinical and Laboratory Standards - CLSI (2019) [9] at the Department of Microbiology - University Medical Center, Ho Chi Minh City, Vietnam from 10/2019 to 04/2020.
Research on applying carbapenemase inactivation method (mCIM) according to CLSI in 2019. Principle: Carbapenemase in bacterial suspension was tested with hydrolyze meropenem plates. The meropenem dish was tested for susceptibility on agar culture plates E. coli ATCC 25922, the strain sensitive to meropenem. There are 4 common antibiotics in carbapenem, however, ertapenem doesn't have the spectrum of activity against P. aeruginosa and doripenem isn't common to use in clinical, imipenem is an unstable compound. Therefore, in this study we choose meropenem for the mCIM phenotype and also follow the recommendation of CLSI 2019.
The carbapenemase enzyme was detected when the sterile ring diameter for E. coli was within the specified limits, indicating that the meropenem was inactivated, unable to inhibit E. coli ATCC 25922. Testing with P. aeruginosa for sensitivity > 97% and 100% specificity for detection of genes blaKPC, blaNDM, blaVIM, blaIMP, blaIMI, and blaOXA-type producing carbapenemase.
This procedure describes a novel method, CBDE according to CLSI in 2020, studied by UCLA and Johns Hopkins, to determine the invitro susceptibility of P. aeruginosa, A. baumannii, and Enterobacteriaceae to colistin.
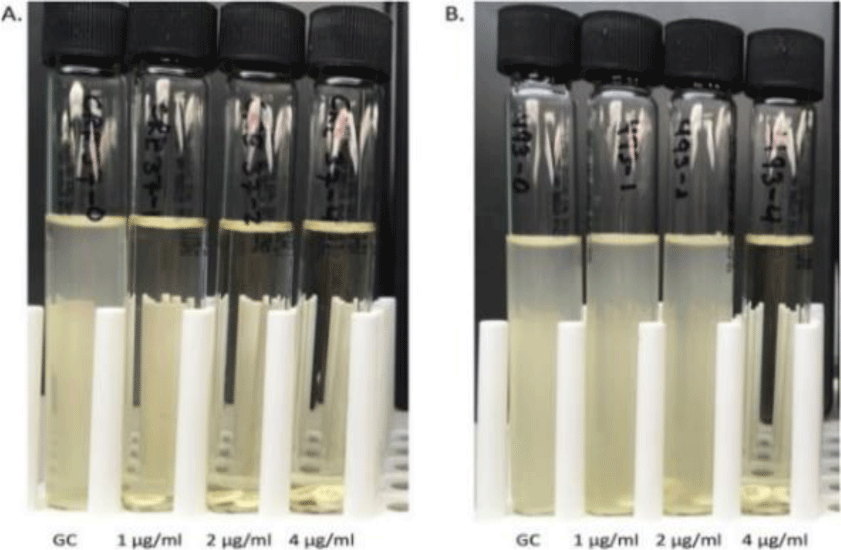
Colistin broth disk elution method (CBDE) was performed with four 10 mL cation-adjusted Mueller-Hinton broth (CA-MHB; Remel, Lenexa) tubes per isolate, to which 0, 1, 2, and 4 colistin disks (10?μg; BD, Sparks MD) were added, generating final concentrations of 0 (growth control), 1, 2, and 4?μg/mL. The tubes were incubated at room temperature for 30 min to allow colistin to elute from the disks. Inocula were prepared by suspending fresh colonies from an overnight sheep’s blood agar plate in normal saline and standardizing the turbidity to match that of a McFarland 0.5 standard. A 50μL aliquot of the standardized suspension was added to each tube and the tubes were gently vortexed for a final inoculum of 7.5×105 CFU/mL, consistent with CLSI 2020 guidelines. Colistin MIC values were read visually, after a 16h to 20h incubation at 35°C in ambient air [10].
The method is performed at the Quality Control Center for Medical Laboratory under Ministry of Health at University of Medicine and Pharmacy at Ho Chi Minh City, Vietnam. Real-time PCR was implemented to determine the presence of blaKPC, blaNDM, blaIMP, blaVIM, blaOXA-48 gene by 2 kit reaction. Amplifications were performed in 25μL reaction volume containing 10μL PCR-mix-1-FRT KPC/OXA-48 or VIM/IMP/NDM, 5μL RT-PCR-mix-mix-2, 0,5μL polymerase and 10μL DNA template. The PCR run was performed using the Sacycler-96 instrument (Sacace biotechnology, Italy). The real-time PCR conditions were as follows: 95°C: 15 min, 5 cycles of 95°C: 5s, 60°C: 20s, 72°C: 15s, 40 cycles of 95°C: 5s, 60°C: 30s, 72°C: 15s. The Sacycler-96 instrument automatically calculated the derivatives of fluorescence measured at 470/525nm: (FAM/Green); 532/570nm (HEX/JOX/Yellow), 585/633nm (ROX/Tesared/Orange), 633/670nm (Cy5/Red), 690/750nm (Cy5.5 Crimson/Quasar 705).
3. RESULTS
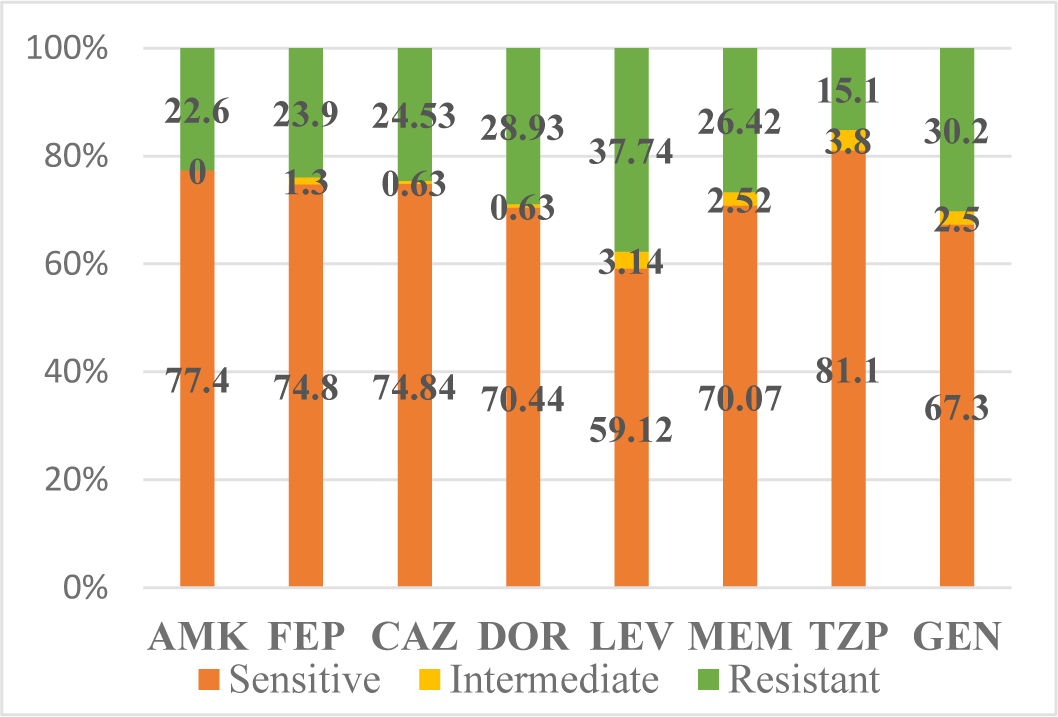
Among 159 strains of P. aeruginosa surveyed, the level of resistance to antibiotics is as follows: amikacin (AMK): 22.6%, cefepime (FEP): 23.9%, ceftazidime (CAZ): 24.53%, doripenem (DOR): 28.93%, levofloxacin (LEV): 37.74%, meropenem (MEM): 26.42%, piperacillin/tazobactam (TZP): 15.1%, gentamicin (GEN): 30.2%.
Of the 8 antibiotics commonly used, P. aeruginosa is resistant to two antibiotics above 30%, levofloxacin (LEV), and gentamicin (GEN) of which the highest resistance is levofloxacin (LEV): 37.74%.
Among 46 strains resistant/intermediate to meropenem, 25 strains (54.3%) were positive mCIM testing and all were resistant to meropenem, 21 strains (45.7%) were negative mCIM testing, in which there are 4 intermediate strains and 17 strains resistant to meropenem.
| mCIM | Frequency | Percentages % |
|---|---|---|
| Positive | 25 | 54.3 |
| Negative | 21 | 45.7 |
| Total | 46 | 100 |
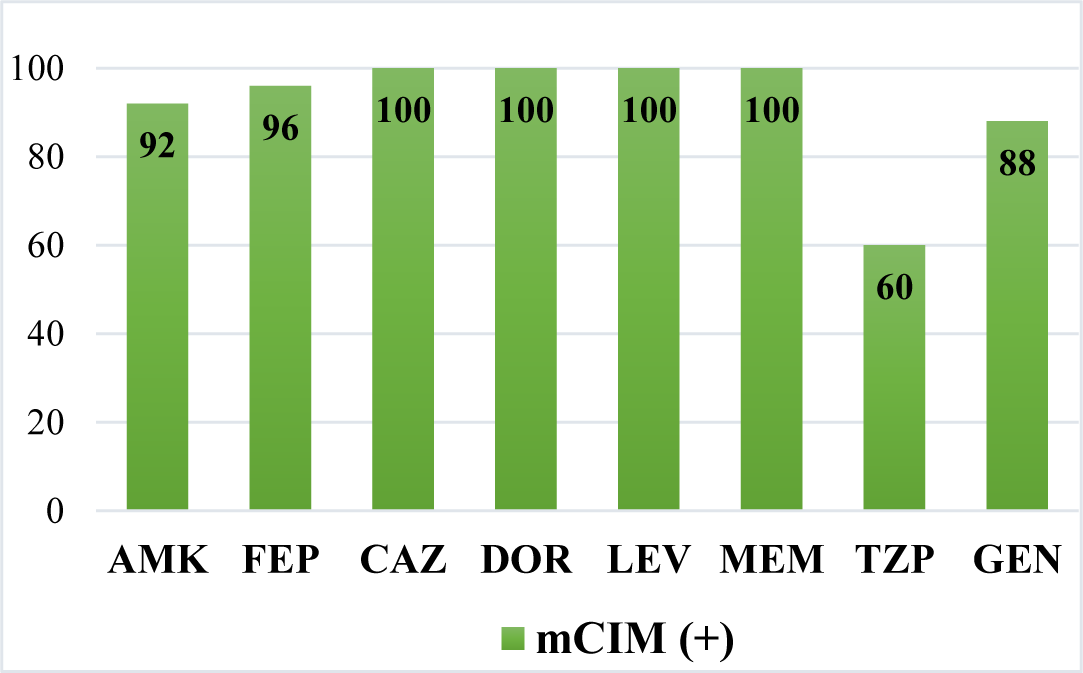
Out of a total of 25 strains (54.3%) P. aeruginosa with carbapenemase producing phenotype mCIM (+) showed resistance to the following antibiotics: amikacin: 92%, cefepime: 96%, ceftazidime: 100%, doripenem: 100%, levofloxacin: 100%, meropenem: 100%, piperacillin/tazobactam: 60%, gentamicin: 88%.
All 8 antibiotics commonly used above, P. aeruginosa are resistant at levels above 80%. In which there are 4 antibiotics 100% resistant to be ceftazidime, doripenem, levofloxacin, meropenem. The only antibiotic with the lowest level was piperacillin/tazobactam: 60%.
In the colistin broth disk elution test phenotype all results are <1 μg/mL. That mean all testing isolates are susceptible to colistin.
The genes with the highest frequency were blaIMP and blaNDM with 21 strains, the lowest was blaVIM, blaKPC with 1 strain.
Number of strains carrying a gene: blaIMP: 4 strains (16%), blaNDM: 2 strains (8%).
Number of strains carrying two genes: blaIMP + blaNDM: 10 strains (40%), blaVIM + blaNDM: 1 strain (4%), blaNDM + blaOXA-48: 1 strain (4%).
Number of strains carrying three genes: blaIMP + blaNDM + blaOXA-48: 6 strains (24%), blaKPC + blaIMP + blaNDM: 1 strain (4%).
4. DISCUSSION
The results revealed that blaIMP was the most prevalence among carbapenem resistance P. aeruginosa. This finding was similar to the result of previous study finding blaIMP on P. aeruginnosa strains [11]. Our results were also concordance with a report of Truong Thien Phu [12] comprised blaIMP: 45%, blaNDM: 11%, blaVIM: 5%, both blaIMP + blaNDM: 5% (n=64). Furthermore, blaNDM, blaKPC and blaOXA-48 which are usually found in Enterobacteriaceae were also detected with the high rate in P. aeruginosa [13-14]. This demonstrates that the ability to acquire resistance genes among bacteria species and the capability to carry multiple antibiotic resistance genes in P. aeruginosa.
The study of colistin-resistant phenotype on P. aeruginosa strains all yielded <1 μg/mL, therefore in this study, MCR genotyping was not performed. Currently, in hospitals in Vietnam, there is no report of resistance to bacteria when using colistin in combination with carbapenem for P. aeruginosa. Meanwhile, the detection of the first plasmid-mediated colistin resistance mechanism regulated by the MCR-1 (mobilized colistin resistance) gene, in 2015 was identified on plasmids from strains of Escherichia coli and Klebsiella pneumoniae on a farm in China [15]. Therefore, in the future, with the spread of resistance genes from animals to humans as well as the horizontal genetic inheritance between different strains of bacteria, it is of great concern in controlling P. aeruginosa strains.
In a study by the authors Khosravi, Yalda Tay, Sun Tee, Vadivelu, Jamuna (2010) [16], at a hospital in Malaysia, the study performed over 90 imipenem-resistant P. aeruginosa cases, 35.5% are carriers of the MβL group, of which 56.2% carry blaVIM and 43.7% blaIMP. According to another study by the authors Yousef Erfani, Sajad Yaghobi, Fatemed Fallah at the hospital in Tehran (2017) [17], the study performed on 212 strains of which A. baumannii (n = 107) and P. aeruginosa (n = 105) resistant and mediated strains to imipenem and meropenem. The results showed that 93 strains out of 105 strains (88.6%) of P. aeruginosa produced enzyme by combination plate test (CDDT) [18], then performed PCR technique, 69 strains (65,7%) carried blaVIM, 36 strains (34.3%) were blaIMP, foundn’t blaNDM-1 in P. aeruginosa. This shows that the prevalence of gene encoding carbapenem over different years and also results from different countries geographically. Therefore, study duration and geographic area can be considered as a contributing factor to these differences.
Therefore, the first step to perform rapid phenotypic screening is a method to select antibiotic-resistant strains of P. aeruginosa according to a fast, simple carbapenemase enzyme production mechanism with high sensitivity and specificity as standard CLSI recommends 2019 and currently follows version 1/2020. Then, accurately determining genotype by multiplex real-time PCR technique helps clinicians orient the selection of antibiotics appropriately, reduce mortality, save costs for patients, as well as prevent and infection control, preventing the spread of these resistance genes.
Limitation of study is the lack of details of patients hospitalizations, outcome and antibiotics received. We did not collect information on the details of the patient admitted to the hospital, when the patient was check out from the hospital, as well as the patient's status on antibiotic treatment. We will look at this issue and further studies will attention.
Conclusion
The inactivation method (mCIM) shows that more than 50% of P. aeruginosa (54.3%) is according to the secretion mechanism carbapenemase enzyme and antibiotic resistance very highly. In the colistin plate elution test, all results were <1 μg/mL. Therefore, colistin is still the salvation option for bacteria resistant to carbapenem. The frequency of occurrence of blaKPC, blaIMP, blaVIM, blaNDM, blaOXA-48 indicates that the acquisition of among bacteria P. aeruginosa’s ability to carry multiple resistance genes causing difficulties in treatment. Method of inactivation of carbapenemase (mCIM), colistin broth disk elution test (CBDE) and the implementation of mutiplex real-time PCR technique can be applied in the Department of Microbiology in Vietnam to support treatment and management antibiotics, infection control, epidemiological surveillance and prevention of the spread of resistant bacteria to the community.