1. INTRODUCTION
Cancer remains one of the most significant global health challenges and the second leading cause of death, with over 19 million new cases and 9.7 million cancer-related deaths reported annually [1]. In Vietnam, cancer is the second leading cause of mortality, following cardiovascular diseases [2]. Despite advancements in treatment, cytotoxic drugs remain central to chemotherapy; however, their non-specific mechanisms often lead to adverse effects in healthy cells and tissues [3].
Recent advances in cancer biology, particularly in understanding oncogenes and their mechanisms, have driven the development of targeted therapies. Among them, cytotoxic drugs that disrupt microtubule function, such as taxanes and vinca alkaloids, have demonstrated notable efficacy. However, their clinical application is often restricted because of resistance and toxicity issues [4]. In the 1990s, a new class of antitumor agents named epothilone was discovered in the myxobacterium Sorangium cellulosum [5]. Epothilones, with their macrolide structure, inhibit cancer cell division by targeting functional microtubules [6]. Notably, they exhibit 1,000 to 5,000 times the potency of Taxol, with significantly reduced side effects. Additionally, analogs of epothilone B have been reported to be up to 35,000 times more potent than Taxol against multidrug-resistant cell lines, highlighting their promise in cancer treatment [7].
The first epothilone was isolated from the S. cellulosum strain So ce90 in 1985 at the Gesellschaft für Biotechnologische Forschung in Braunschweig, Germany, and it exhibited selective antifungal activity against Zygomycete and Mucor hiemalis when cultured in liquid medium [8]. Myxobacteria, typically found in soils rich in decaying vegetation and herbivorous feces, can degrade crystalline cellulose and are cultured using filter paper as the sole carbon source in mineral-salt agar with an inorganic nitrogen source, such as KNO3 or (NH4)2SO4. These bacteria are prolific producers of biologically active secondary metabolites, including epothilone, which has been isolated from diverse S. cellulosum strains globally [9,10].
Despite this potential, limited research in Vietnam has examined myxobacteria for their antifungal and antitumor properties, particularly those related to epothilone. This study identified myxobacterial strains from Vietnam capable of producing novel bioactive compounds with antifungal and antitumor activities.
2. MATERIALS AND METHODS
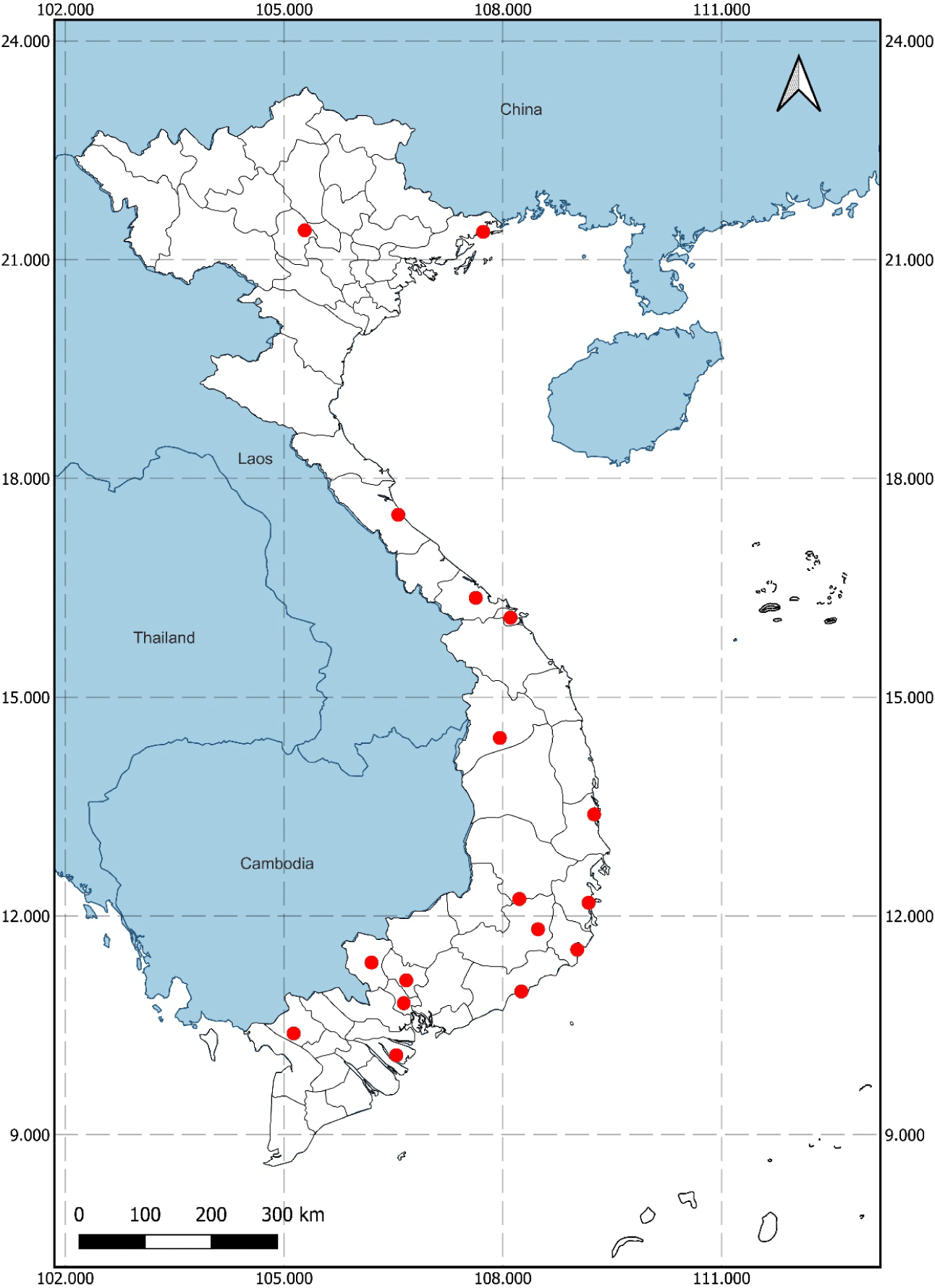
Following established protocols for isolating cellulose-degrading microorganisms from humid, high-temperature environments with high organic carbon contents (e.g., decaying wood and haystacks), 41 soil samples were collected from various provinces and cities in Vietnam (Fig. 1). Each sample was labeled with a unique code consisting of the first two letters of the sampling site and a two-digit ordinal number. Samples were stored in sterile 50 mL Falcon tubes (Thermo Fisher Scientific, Waltham, MA, USA; catalog number: 339653), with each tube containing 25% to 50% soil.
Strains of dermatophytes, molds, and cell lines were obtained from the Department of Microbiology - Parasitology, Faculty of Pharmacy, University of Medicine and Pharmacy at Ho Chi Minh City, Vietnam. These included Trichophyton mentagrophytes, Microsporum gypseum, Trichophyton rubrum, Aspergillus fumigatus, Aspergillus flavus, Aspergillus niger, Penicillium sp., Fusarium sp., and Mucor sp. The cell lines used were breast cancer cells (MCF-7), human embryonic kidney cells (HEK-293), and human liver carcinoma cells (HepG2).
Myxobacterial strains were isolated using the following four-step process:
Step 1: Macroscopic examination
Bacteria were initially cultured on CY agar (containing 0.3% casitone, 0.1% yeast extract, 0.1% CaCl2.2H2O, 1.5% agar, and pH adjusted to 7.2) and VY/2 agar (containing 0.5% bakers’ yeast, 0.1% CaCl2.2H2O, 0.5 mg/µL cyanocobalamin, 1.5% agar, and pH adjusted to 7.2) to observe colony characteristics. Cultures were prepared in triplicate to ensure consistency.
Step 2: Microscopic observation
Gram staining was conducted using a Gram staining kit (Sigma-Aldrich, St. Louis, MO, USA; catalog number: 77730), followed by microscopic analysis of bacterial morphology. Each observation was repeated three times independently to confirm reproducibility.
Step 3: Cellulose degradation assessment
Strains were screened for cellulose degradation in carboxy methyl cellulose (CMC) medium (containing 1 g K2HPO4, 1 g(NH4)2SO4, 0.5 g MgSO4.7H2O, 0.001 g NaCl, 10 g CMC, and 500 mL H2O) as the sole carbon source. This screening was performed in three independent trials to confirm consistent cellulose degradation.
Step 4: Fruiting body formation
The formation of fruiting bodies was assessed using the VY/2 medium, and the tests were repeated three times to ensure reproducibility.
For cellulose degradation assessment, 5 g of dry soil was dispersed in 20 mL of 0.85% NaCl solution, allowed to settle for 5 min, and diluted to 10–1, 10–2, and 10–3. A 100 µL aliquot of the diluted soil suspension was plated on CY medium and incubated at 30°C for 3 days. Colonies with yellow-orange, red, or brown hues were transferred to VY/2 agar and incubated at 30°C for 3–10 days. Colonies exhibiting orange or brown fruiting bodies or radial growth were Gram-stained. Strains presenting as Gram-negative bacilli with round or square heads (0.8–1.2×3–8 µm) and spore-like structures (2–4 µm) were selected, purified through repeated inoculation on VY/2 medium, and stored on sterile filter paper (Whatman, Maidstone, UK; catalog number: 1001090).
To assess motility, VY/2 agar was spread thinly on a glass slide and solidified. A fruiting body was inoculated onto the slide, sealed with a 1:1 mixture of vaseline and paraffin, and incubated at 30°C for 3–10 days. Fruiting bodies were collected using a sterile toothpick and were transferred to CMC medium. Bacillus subtilis PY79 served as the positive control for cellulose degradation, whereas Escherichia coli served as the negative control. After 4–5 days of incubation at 30°C, cellulose degradation was detected using Congo red staining (0.1% w/v; Sigma-Aldrich; catalog number: C6767) and NaCl (1M; Sigma-Aldrich; catalog number: S9888). A clear zone around the colony indicates positive cellulose degradation. All degradation tests were conducted in triplicate to ensure consistent results.
Primary bacterial cultures were initially grown in M26 medium (containing 0.8% potato starch, 0.2% glucose, 0.2% tryptone, 0.2% yeast extract powder, 0.1% CaCl2.2H2O, 1 mL/L trace elements solution, 1% agar and pH adjusted to 7.0) at 200×g for 3 days. The strains were then transferred to 100 mL of CK6 medium (containing 0.15% MgSO4.7H20, 0.002% Fe3+ citrate, 0.2% KNO3, 0.025 K2HPO4, 0.5% glucose, and 0.15% CaCl2.2H2O) supplemented with 0.72 g of Amberlite XAD-16 resin (MilliporeSigma Supelco, Burlington, USA; Cat. No. XAD16500G), cultured at 200×g, and incubated at 30°C for 10 days. After incubation, the resin was separated, rinsed with water, and dried in a desiccator.
The dried resin was eluted with 50 mL of methanol (Merck KGaA; catalog number: 106009) and then evaporated under vacuum at 40°C. The residue was dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich; catalog number: D8418) for use in antifungal and cytotoxicity assays and in 1 mL of methanol for thin-layer chromatography (TLC) analysis. All cultivation and extraction procedures were performed in triplicate to ensure consistency.
Myxobacterial extracts were analyzed by TLC on silica gel Si 60 F254 plates (Merck KGaA; catalog number: 105554). The mobile phase was a mixture of dichloromethane (Merck KGaA; catalog number: 106050) to methanol (Merck KGaA; catalog number: 106009) in a 90:10 ratio. Potential bioactive compounds were detected by UV irradiation at 254 nm. Each TLC analysis was performed in triplicate to ensure consistent detection of bioactive compounds.
Dermatophytes and molds were cultured on SDA agar slants (HiMedia Laboratories; catalog number: MH063) at 30°C for 3–7 days, depending on the species. Fungal suspensions were adjusted to an optical density (OD) of 0.1 at 530 nm, corresponding to 1–5×106 CFU/mL. The suspension was spread onto agar plates, and 6-mm-thick wells were drilled under sterile conditions. Crude extracts, dissolved in DMSO to a final concentration of 20.48 mg/mL, were added at 60 µL per well. The plates were then incubated at 30°C for 3–7 days, with antifungal activity indicated by an inhibition zone around each well. Ketoconazole (HiMedia Laboratories; catalog number: CMS4322) was used as a positive control to validate the antifungal activity of the myxobacterial extracts. Each antifungal assay was conducted in triplicate to ensure reproducibility.
Additionally, antifungal activity was assessed using TLC bioautography. TLC plates were placed in contact with agar plates inoculated with fungal suspensions and incubated at 30°C for 3–7 days. Inhibition zones on the agar, corresponding to spots on the TLC plate, indicated antifungal activity. All tests were performed in triplicate to confirm the consistent results.
Cytotoxicity was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay (Sigma-Aldrich; catalog number: M2128). MCF-7, HEK-293, and HepG2 cells were cultured in MEM medium (Sigma-Aldrich; catalog number: M2279) with 10% fetal calf serum (FCS; Sigma-Aldrich; catalog number: F4135) at 37°C for 24 h in 96-well microtiter plates (Thermo Fisher Scientific; catalog number: 167008). The MTT reagent was prepared at concentrations ranging from 50 ng/mL to 6400 ng/mL in MEM with 10% FCS. After 24 h of incubation, 10 µL of 5 mg/mL MTT solution was added to each well, followed by an additional 4 h of incubation. The culture medium was then removed, and 0.2 mL of DMSO was added to dissolve the formazan crystals. Absorbance was measured at 570 nm using a microplate reader (Bio-Rad, Hercules, CA, USA; model: iMark). The assay was conducted in triplicate, and IC50 values were calculated using GraphPad Prism v5.04 (GraphPad Software, San Diego, CA, USA). The percentage of cell inhibition was calculated as inhibition = [(OD_control – OD_test) / OD_control] × 100.
3. RESULTS
From 41 soil samples collected across various provinces in Vietnam, 33 strains were isolated, exhibiting colonies with yellow, orange-yellow, brown, or black-brown pigmentation on CY and VY/2 media. These strains included AG01, AG02, AG03, BD01, BD02, BD03, BD04, BD05, BT01, DL01, DL02, DL03, DL04, DL05, DN01, H02, H03, HCM01, HCM03, KH03, KT01, LD02, LD03, PR01, PR02, QB01, QN01, QN02, QN03, QN04, QN05, TN01, and TN02. Of these, six strains (AG01, BD04, H02, QN01, QN02, and QN04) demonstrated cellulose degradation activity CMC medium. Additionally, 17 strains were identified as Gram-negative bacilli, and three strains (QN01, QN02, and QN04) formed fruiting bodies. The detailed results are presented in Table 1 and Figs. 2–4.
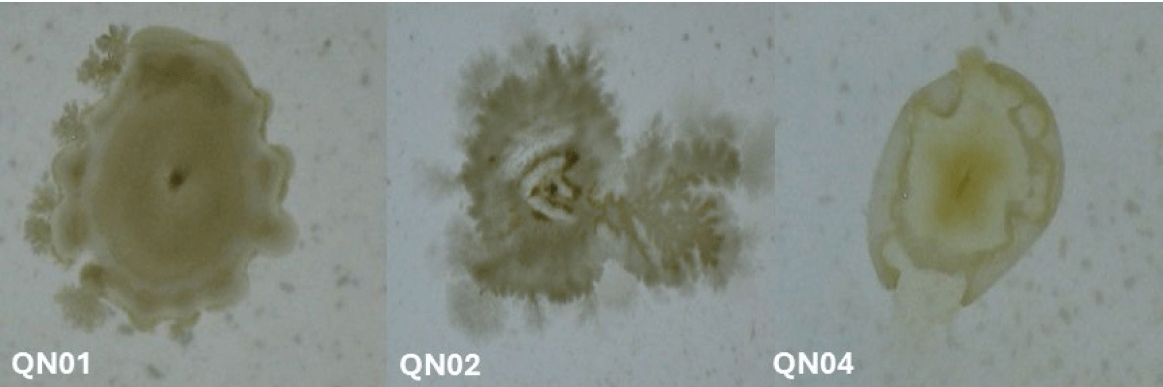
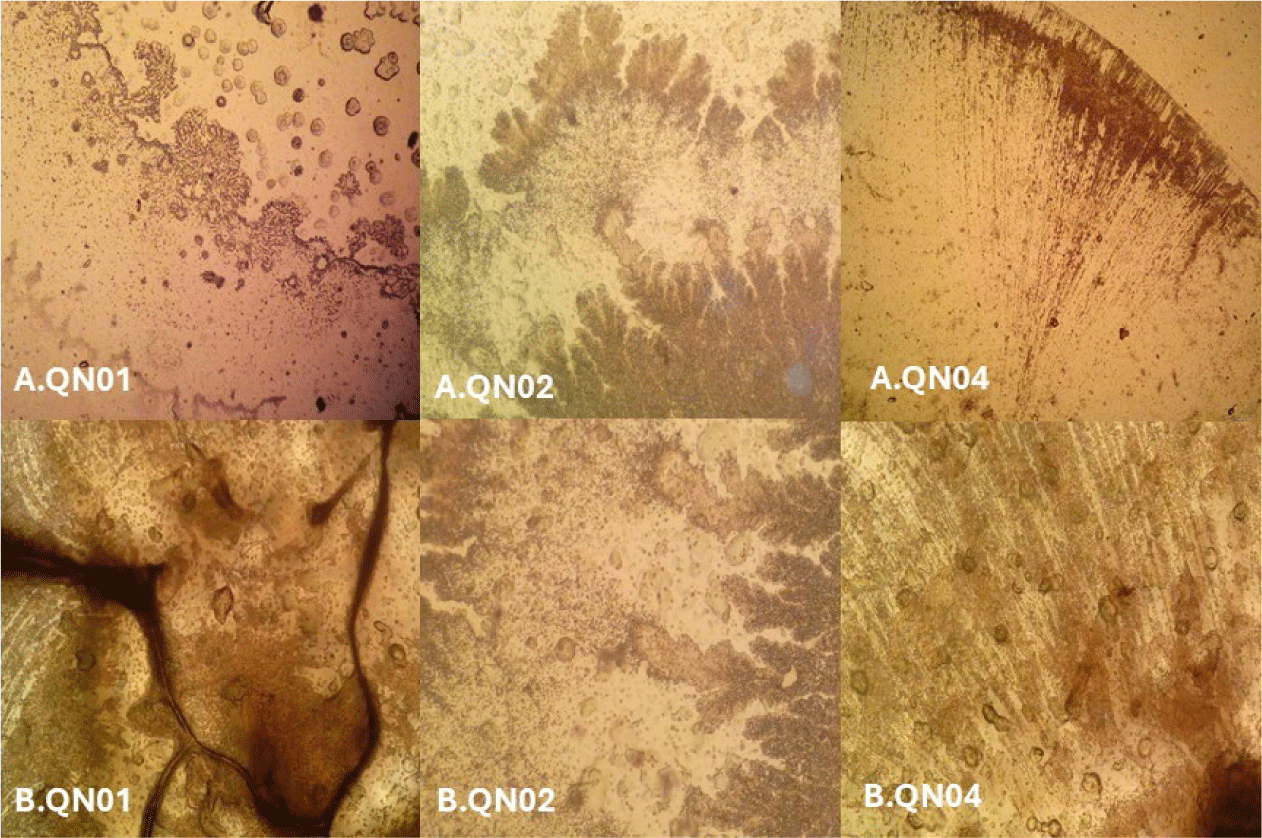
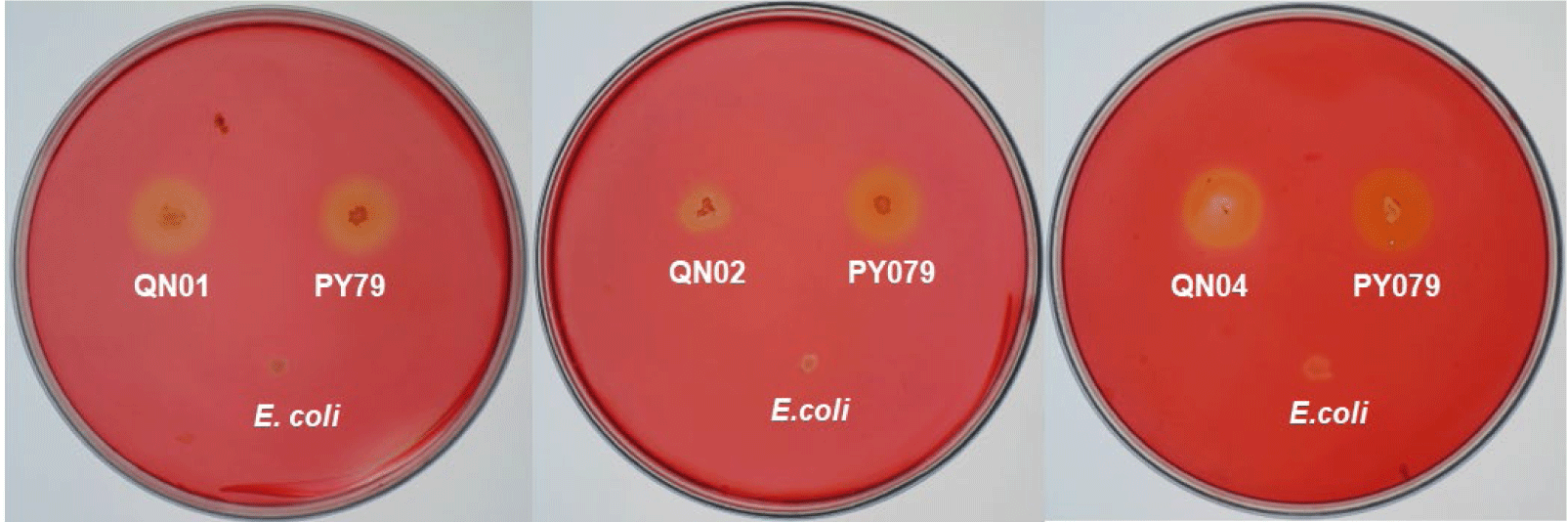
Macroscopic and microscopic examinations revealed that QN01, QN02, and QN04 exhibited characteristics consistent with S. cellulosum, according to Bergey’s Manual of Systematic Bacteriology. These strains, isolated from soil from Quang Nam province, demonstrated cellulose degradation, as confirmed by comparing the clear zones around the colonies with those of the positive control strain B. subtilis PY79 and the negative control strain E. coli. Their degradation capabilities confirmed their identification as S. cellulosum.
Morphological analysis of VY/2 yeast agar revealed distinct features for each strain. QN01 displayed yellow-brown colonies with undulating edges, dark veins, and scattered spherical fruiting bodies near the colony boundary. QN02 initially formed milky-white, round colonies with serrated edges and concentric rings. After 5–7 days, the colonies developed branched vein-like structures and brown spots with thin, brittle patches. Under 150× magnification, brown sporocarps with oval or polygonal shapes were observed. QN04 showed yellow-brown colonies with rounded margins and radial pocket-shaped structures, with visible oval-shaped fruiting bodies at 150× magnification. All three strains exhibited large fruiting bodies (30–50 µm), Gram-negative bacilli with slightly round heads, and spores with gliding motility, consistent with myxobacterial features.
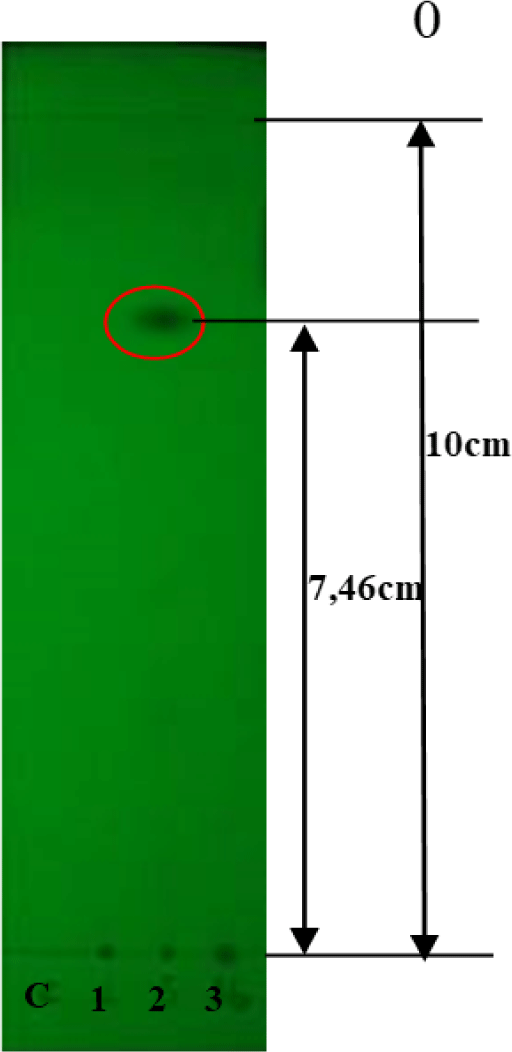
TLC analysis of the crude extracts indicated that only QN02 exhibited a UV absorption spot at 254 nm with an Rf value of 0.746 (Fig. 5). Visualization with vanillin or 1% concentrated H2SO4 revealed three distinct spots on the chromatogram, with Rf values of 0.5, 0.7, and 0.746.
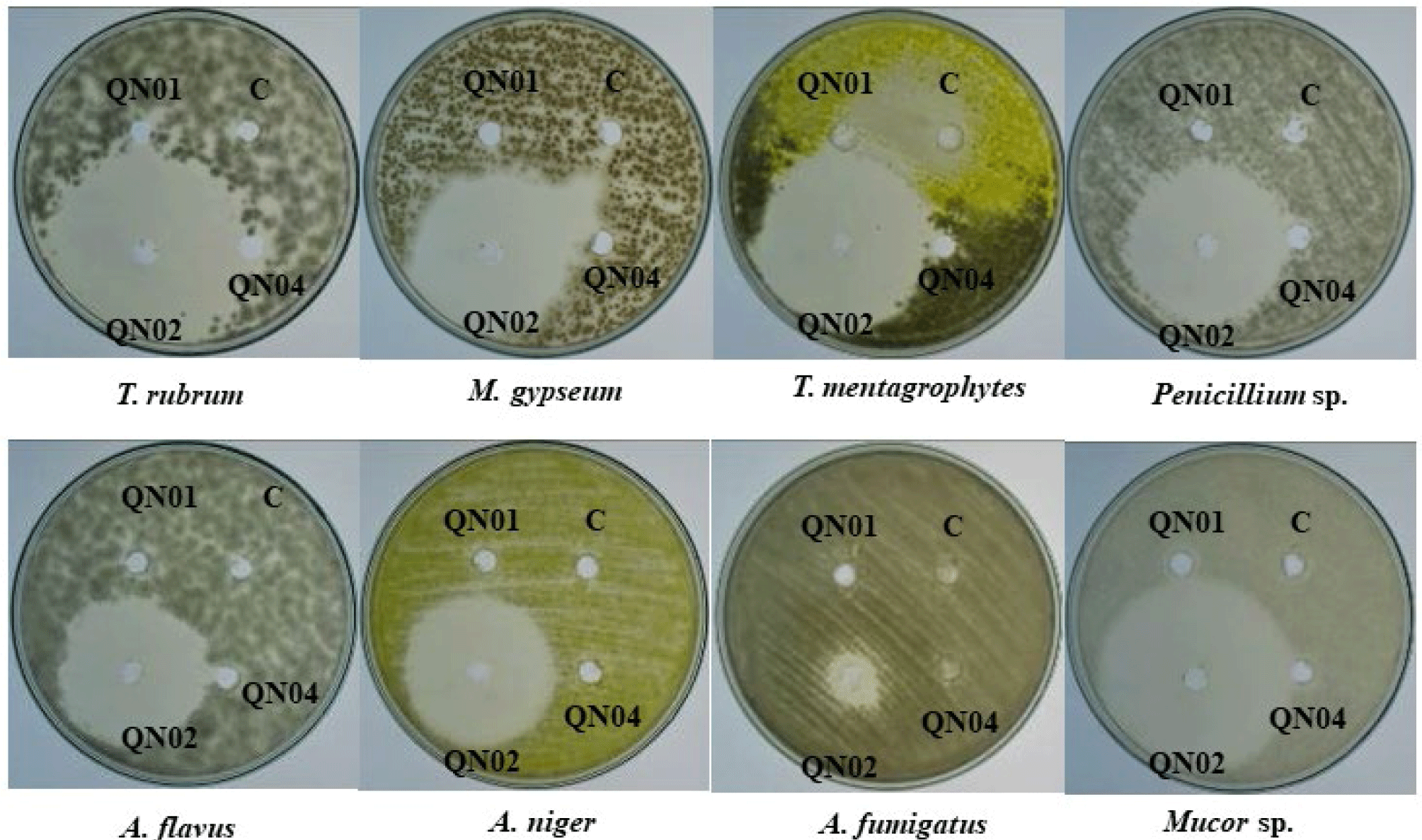
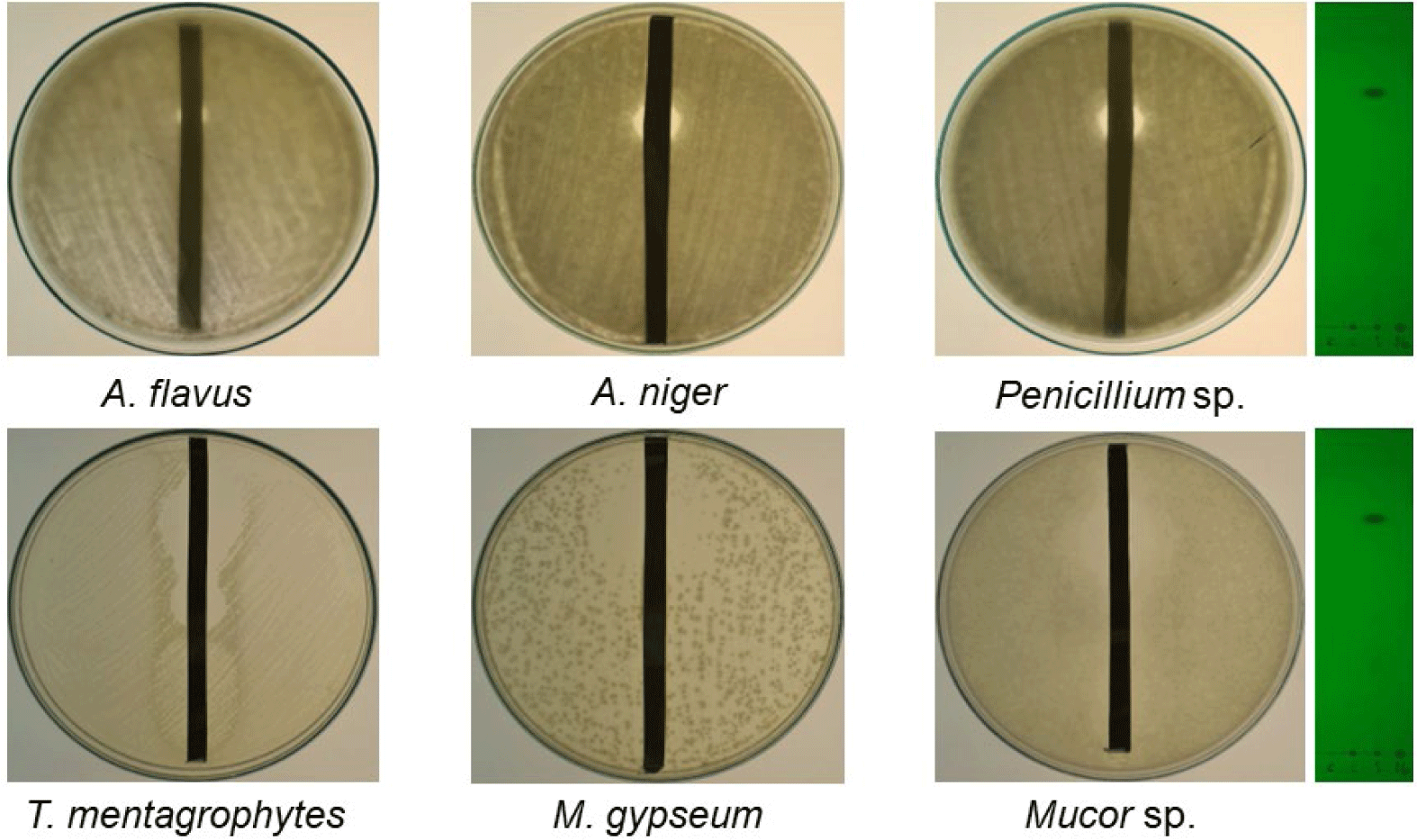
The antifungal and antibacterial activities of the crude extracts of QN01, QN02, and QN04 were evaluated using the disk diffusion method, and the results are summarized in Tables 2 and 3, and Figs. 6 and 7. The QN02 strain showed significant antifungal activity against all tested fungi, except for Candida albicans and Fusarium sp., with inhibition zones greater than 15 mm. TLC bioautography further confirmed the antifungal activity of the QN02 extract, particularly at Rf=0.746, against several fungi, including T. mentagrophytes, M. gypseum, Mucor sp., Penicillium sp., A. flavus, and A. niger. The consistent antifungal activity at Rf=0.746 indicates that QN02 contains a potent bioactive compounds, making it a strong candidate for further cytotoxicity testing.
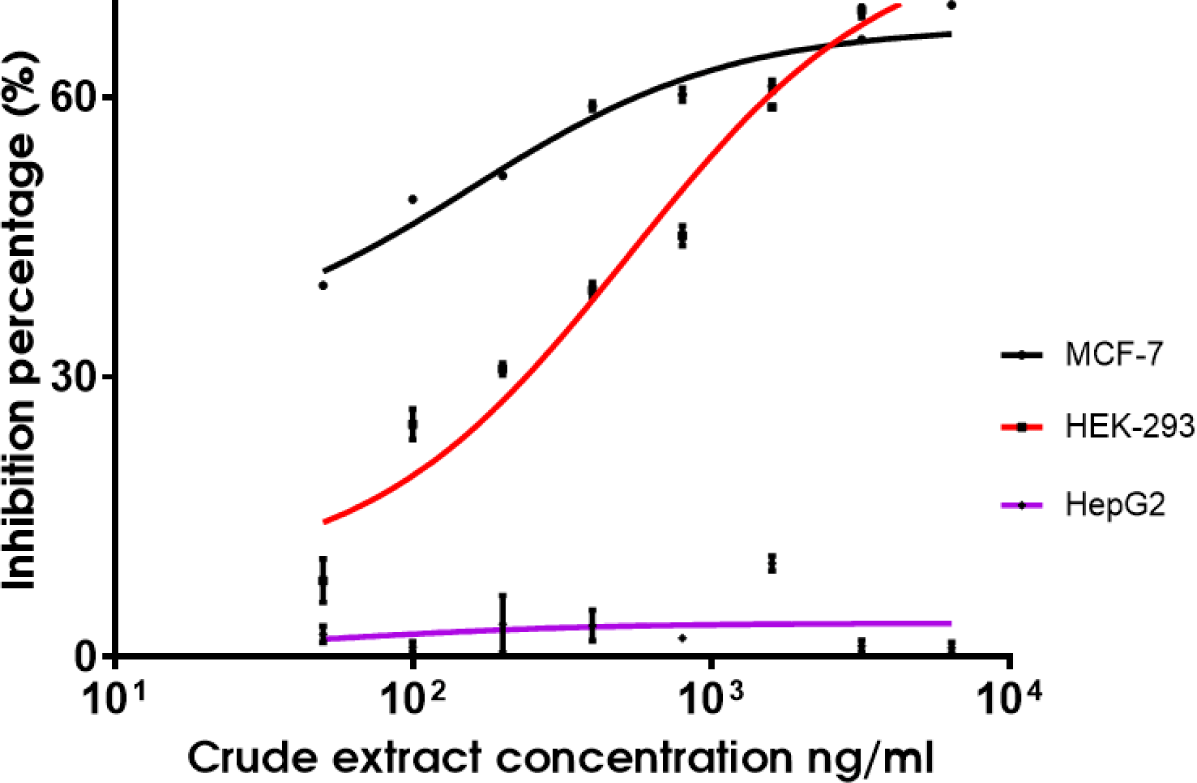
The cytotoxicity of the QN02 crude extract was evaluated using the MTT assay. Results indicated a dose-dependent decrease in absorbance at 570 nm, correlating with reduced cell viability as the extract concentration increased (Table 4, Fig. 8). The IC50 values of the QN02 extract were 0.16 µg/mL in MCF-7 and 0.53 µg/mL in human kidney cells (HEK-293). However, the extract did not inhibit the proliferation of HepG2 cells at concentrations up to 6.4 µg/mL.
4. DISCUSSION
The isolation and characterization of myxobacteria from 41 soil samples collected across 15 provinces in Vietnam led to the identification of three strains, designated QN01, QN02, and QN04, which meet the taxonomic criteria for S. cellulosum as outlined in Bergey’s Manual of Systematic Bacteriology. All three strains were derived from soil samples collected in Quang Nam province. The geographic distribution of myxobacteria detected across multiple provinces highlights their widespread presence in diverse environments. This information is crucial not only for understanding environmental preferences and potential regional adaptations but also for guiding future researchers in exploring myxobacteria in similar or neighboring areas.
This mapping of sample sites (Fig. 1) provide a foundation for identifying regions with high myxobacterial diversity, which could lead to novel bioactive compounds. The identification of these strains aligns with previous studies by Gerth et al. and Hyun et al., who isolated S. cellulosum using filter paper or CMC as the sole carbon source [7,10,11].
Our findings suggest that the QN02 strain is a promising producer of epothilone, based on its antifungal and cytotoxic activities. TLC analysis of the QN02 extracts revealed three distinct spots with Rf values of 0.5, 0.7, and 0.746, which align with with the separation of epothilone A and B as observed in studies by Gerth et al. [7]. The solvent system (dichloromethane-methanol, 90:10) effectively separated these compounds, supporting the presence of epothilone in the QN02 extract.
In our cytotoxicity assays, the QN02 extract had an IC50 of 0.16 µg/mL against the MCF-7 breast cancer cell line, indicating potent antiproliferative activity. Although Kamath et al.reported an IC50 of 3.5 nM for purified epothilone, this result is consistent with the IC50 values observed in other studies on crude myxobacterial extracts [12]. For instance, Wang et al. reported an IC50 of 1.32 µg/mL and Guo et al. found an IC50 of 1.07 µg/mL for similar methanol extracts, suggesting that the crude form retains significant cytotoxic activity [13,14].
Comparative studies have demonstrated that epothilone exhibits superior cytotoxicity compared with Taxol across various cancer cell lines, with IC50 values ranging from 0.3 to 2.0 nM, whereas Taxol IC50 values vary significantly from 2 to 9,000 nM, depending on the cell type [8]. These differences in cytotoxicity emphasize the influence of extraction methods and the varying sensitivities of cancer cell lines to epothilone.
Despite these promising results, this study has several limitations. The sample size limited to 41 soil samples from various specific regions in Vietnam may not represent the full diversity of cellulose-degrading myxobacteria. Additionally, further molecular characterization, including DNA sequencing, is required to confirm the taxonomy of the isolated strains and to elucidate the biosynthetic pathways responsible for producing bioactive compounds.
5. CONCLUSION
This study confirmed the presence of cellulose-degrading myxobacteria in soil, reinformcing their abundance in specific environments, which aligns with previous research findings. Among the isolated strains, QN02 showed significant potential for epothilone production, highlighting its promise as a source of bioactive compounds.