1. INTRODUCTION
Pseudomonas aeruginosa, a ubiquitous Gram-negative bacterium, thrives in diverse environments including soil, aqueous surfaces [1,2], as well as multiple human body sites such as the skin, throat, and gastrointestinal tract [3,4]. Its remarkable adaptability and potent antibiotic resistance enable this bacterium to survive and colonize a wide range of healthcare settings, making it one of the most common nosocomial pathogens in clinical setings [5-7]. P. aeruginosa infections frequently occur as opportunitic infections, posing a particularly grave threat to immunocompromised individuals. The spectrum of diseases attributed to P. aeruginosa ranges from milder, atypical infections like ear infections and skin rashes to more severe and complex conditions, such as burn infections, cellulitis, necrotizing fasciitis, pneumonia (especially ventilator-associated pneumonia), sepsis, and toxic shock syndrome, among others.
The primary cause leading to treatment failure in P. aeruginosa infections is the high and escalating level of antibiotic resistance in clinical settings. P. aeruginosa, classified as a multidrug-resistant bacterium, ranks among the most dangerous and urgent pathogens in the search for new antibiotics, according to the CDC and WHO [8,9]. The elevated antibiotic resistance in P. aeruginosa is attributed to the combination of various mechanisms, including (1) low permeability and reduced cellular uptake of antibiotics, (2) overexpression of antibiotic efflux pump systems, (3) modification of antibiotic target sites, and (4) secretion of degrading enzymes or alteration of antibiotic structures. These resistance mechanisms may arise from inherent resistance capabilities or develop through mutations or acquisition of resistance genes from external sources. Additionally, P. aeruginosa has the ability to modulate gene expression based on growth phases, environmental conditions, or responses to stress factors, contributing to antibiotic resistance at various levels - known as adaptive resistance mechanisms [10,11].
The expression of multidrug resistance efflux pumps plays an important role in the high-level resistance of Gram-negative bacteria, including P. aeruginosa [12]. These pumps are classified into five main families: the ATP-Binding cassette (ABC) family, major facilitator superfamily family, small multidrug resistance family, resistance nodulation division (RND) family, and multidrug and toxic compound extrusion (MATE) family. In P. aeruginosa, the RND efflux pumps are the most prevalent. These complex molecular machinery components are composed of membrane transport proteins (such as Mex B, MexY, MexD, MexF...), which associate with outer membrane porins (OprM, OprJ, OprN…) through linking proteins in the periplasm (MexA, MexX, MexC, MexE…) [13]. Notably, the genome of P. aeruginosa harbors at least 12 distinct RND efflux pumps, among these, MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM have significant clinical relevance. These efflux pumps are typically expressed at low levels or inhibited in vitro. Mutations in regulatory genes controlling their expression, particularly those repressing operons containing genes encoding these pumps, are the primary drivers behind the development of a multidrug resistance phenotype in clinical strains [12-17].
Inhibition of efflux pump systems has gained significant attention in recent years. Efflux pump inhibitors (EPIs) can reverse bacterial resistance to existing antibiotics, reduce toxicity, or inhibit biofilm formation in certain pathogens [18,19]. One of the first EPI studied, Phe-Arg-β-naphthylamide (PaβN), can inhibit the RND efflux pumps of various Gram-negative bacteria by binding to the distal binding site of the efflux pump and hindering the movement of the G-loop, which is essential for efflux activity [20]. However, PaβN is not suitable for clinical use due to its toxicity on eukaryotic cells [21]. Some pyridopyrimidine compounds like ABI-PP (or D13-9001) can bind to the distal binding site of both the AcrB subunit in E. coli and MexB in P. aeruginosa, inhibiting the functional movement of these proteins. However, ABI-PP lacks the ability to inhibit other similar efflux pump systems (such as MexXY in P. aeruginosa), which limits its value in clinical setting [22,23]. This highlights the challenge associated with the research and screening of EPIs due to the diversity among the structural patterns of bacterial efflux pump systems. Therefore, it is crucial to gather clinical data on the expression rates of multidrug-resistant efflux pump systems and their correlation with bacterial resistance patterns, as well as the factors contributing to increased expression of antibiotic efflux pumps, to aid in the development of EPIs in clinical research.
In Vietnam, P. aeruginosa is one of the most closely monitored pathogens regarding antibiotic resistance. However, current domestic studies primarily focus on evaluating resistance levels of this bacterium to various antibiotic classes using in vitro methods like disk diffusion and minimum inhibitory concentration (MIC) determination by dilution. To our knowledge, no report have been piblished aiming to determine the overexpression of efflux pumps in P. aeruginosa.
2. MATERIALS AND METHODS
P. aeruginosa strains from inpatients were collected and isolated using standard procedures at the Microbiology Departments, University Medical Center of Ho Chi Minh City (215 Hong Bang St., Ward 11, District 5), and Le Van Thinh Hospital (130 Le Van Thinh, Binh Trung Tay Ward, Thu Duc City). From May to July 2023, all strains that suspected of P. aeruginosa were picked up and subcultured onto the MacConkey agar. Then, pale colonies that showed positive oxidase test and emitted fluorescence under UV light when cultured on Cetrimide agar (Merck, Darmstadt, Germany) were streaked onto the Luria-Bertani agar. Next, transfer a small amount of bacteria from a single colony of each strain into 20 µL of Q1 and incubate at 90℃ for 5 mins to prepare a cell lysate. One µL of the cell lysate was added into a PCR mixture which consists of 12.5 µL of 2× PCR master mix (Thermo Scientific, Waltham, MA, USA), 1 µL of each primer (10 µM), and Q1 up to a final volume of 25 µL. Two pairs of primers, PA-GS and PA-SS, were used separately to amplify the specific region in the 16S rRNA gene of the Pseudomonas genus and the specific sequence in V2–V8 region of the 16S rRNA gene of P. aeruginosa, respectively [24]. PCR amplification was performed in a SimpliAmp Thermal cycler (Thermo Fisher Scientific) with an initial denaturation step at 95℃ for 5 min, followed by 30 cycles at 95℃ for 30 s, 51.0℃ for 30 s, and 72℃ for 30 s and a final extension step at 72℃ for 10 min. The PCR products were separated by 2% agarose gel electrophoresis. Strains that produced 2 specific products at 618 bp and 956 bp were identified as P. aeruginosa, while other Pseudomonas sp. just produced a product at 618 bp. Clinical P. aeruginosa were stored in 20% glycerol at –80℃ for later experiments. P. aeruginosa ATCC 27583 and E. coli ATCC 25922 were used as positive and negative controls for PCR reactions, respectively (Table 1).
The antibiotic susceptibility of isolated P. aeruginosa strains was determined using the agar disk diffusion and broth microdilution methods as described in the guidelines of CLSI M07-A11 and M100-S33 (Clinical & Laboratory Standard Institue, Malvern, PA, USA) [25,26]. The antipseudomonal antibiotic disks including piperacillin-tazobactam (100 µg/10 µg), ceftazidime (30 µg), meropenem (10 µg), gentamycin (10 µg), and ciprofloxacin (5 µg) (purchased from Nam Khoa Biotech, Ho Chi Minh City, Vietnam) were used for the disk diffusion assay. Whereas, cefepime, imipenem, amikacin, levofloxacin, and colistin (purchased from Merck) were selected for the broth microdilution assay. P. aeruginosa ATCC 27583 was used as control in antibiotic susceptibility assays. Each antibiotic susceptibility assays was performed in triplicated.
Phe-Arg-β-naphthylamid dihydrochloride (PaβN) inhibits the activity of the RND efflux pump family, which in turn can reverse the resistant trait of P. aeruginosa to antibiotics that are substrates of these pumps. A broth microdilution assay was used to determine the MIC of the reporter antibiotics on P. aeruginosa in the presence (MICp) and absence (MICa) of PaβN (50 mg/L) [27] (Table 2). The MIC reduction ratio (MICr) of antibiotics on each P. aeruginosa strain in the presence and absence of PaβN was then calculated as follows: MICr = MICa / MICp. P. aeruginosa ATCC 27583, a susceptible strain, was used in this phenotypic assay for negative control. If the MICr of the corresponding reporter antibiotic on a certain P. aeruginosa strain was higher than the corresponding MICr of the negative control, it indicated the overexpression of that type of efflux pump in that particular strain. The phenotypic asay was performed in triplicate on each strain.
| Antibiotic | Efflux system |
|---|---|
| Carbenicillin | MexAB-OprM |
| Erythromycin | MexCD-OprJ |
| Ofloxacin | MexEF-OprN |
| Gentamicin | MexXY-OprN |
3. RESULTS
From May to July of 2023, a total of 73 samples suspected of P. aeruginosa infection were collected from University Medical Center of Ho Chi Minh City and Le Van Thinh hospital. After being isolated on MacConkey and Cetrimide agar, 63 strains that displayed colorless colonies (on MacConkey agar), were oxidase-positive, and produced fluorescence (on Cetrimide agar) were selected for identification via PCR assays. The results from the PCR tests confirmed that 60 strains were positive for P. aeruginosa. These strains were mostly isolated from respiratory specimens (71.67%; Table 3).
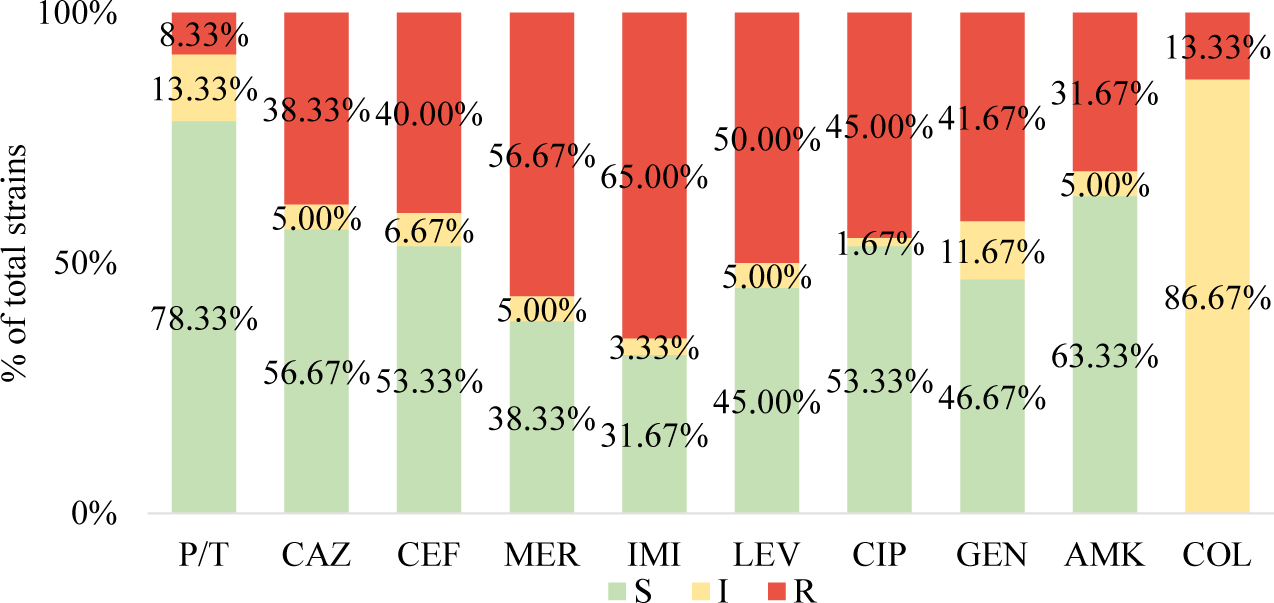
P. aeruginosa isolates exhibited a high resistance rate to the antibiotics tested as shown in Fig. 1. In disk diffusion assays, the highest resistance rates were observed for meropenem (56.67%) and ciprofloxacin (45.00%). Gentamicin and ceftazidime were resisted by these strains at a rate of 41.67% and 38.33%, respectively. However, the combination of piperacillin and tazobactam still showed high susceptibility, with 91.67% of strains being susceptible. The MIC values also revealed high resistance rates of the isolates to imipenem, levofloxacin, cefepim, and amikacin, with resistance rates of 65.00%, 50.00%, 40.00%, and 31.67%, respectively (Table 4). Meanwhile, the efficacy of colistin remained high, with a low resistance rate of 13.33%. Notably, 28 strains (46.67%) were found to be resistant to at least three different classes of antibiotics, classifying them as multi-drug resistant strains.
Out of 60 isolates, 38 strains exhibited overexpression of at least one type of efflux pump, constituting for 63.33%. Among these, MexEF-OprN was the most frequently overexpressed efflux pump system (32/60 strains), representing 53.33%. MexCD-OprJ overexpression was detected in 13/60 strains (21.67%), while MexAB-OprM was overexpressed was detected in 6/60 strains (10.00%). The lowest overexpression was observed for MexXY-OprM, present in 5/60 strains (5.00%). In strains with overexpression of efflux pumps, 13 strains simultaneously exhibited overexpression of two efflux pump systems, accounting for 21.67% of the total isolated strains. MexCD-OprJ and MexEF-OprN were the two most commonly co-expressed efflux pump systems, appearing in 10/60 strains (16.67%) (Tables 5 and 6).
| Type of efflux pump | Number of strains | % of total strains |
|---|---|---|
| MexAB-OprM | 6 | 10.00 |
| MexCD-OprJ | 13 | 21.67 |
| MexEF-OprN | 32 | 53.33 |
| MexXY-OprM | 3 | 5.00 |
Table 7 shows the number of strains that overexpress each type of efflux pump based on the infection site. The overexpression of each efflux pump was controlled by different cellular mechanisms and may not be directly related to each other. Therefore, the relationship between the sampling site and the overexpression rate of each efflux pump was analyzed separately using Fisher’s Exact test in Rstudio software (R 4.3.0). The p-values obtained were greater than the significance level of 0.05 (detailed statistical analysis provided in the supplementary file). This indicates that there is no significant relationship between the two factors.
The number of strains overexpressing efflux pumps and the resistance rates to antibiotics are presented in Table 8. The number of strains overexpressing MexAB-OprM and MexXY-OprM efflux pumps observed in the study is relatively low. Therefore, the analysis focused only on the correlation between overexpression of the MexEF-OprN and MexCD-OprJ systems with antibiotic resistance. An overexpression of a certain type of efflux pump can confer bacteria resistance to different antibiotics. Also, a certain antibiotic could be the substrate to be pumped out by different efflux pumps. Moreover, aisde from efflux pump overexpression, P. aeruginosa could trigger resistance to antibiotics by various mechanisms. These include lowering the penetration of antibiotics into the cell by down-regulating or diminishing the expression of specific porins, weakening the binding of antibiotics to their targets by altering or mutating the antibiotic cellular targets, or modifying or destroying antibiotic structures by producing various enzymes. This complexity cannot be fully analyzed with our limited data. Therefore, in order to determine the possible relationship of the efflux pump overexpression and antibiotic resistance, the Chi-squared test (or Fisher’s exact test when there was count that less than 5) was used to analysed the relationship of each pair of certain efflux pump overexpression and single antibiotic. The statistical tests revealed a significant impact (p<0.05) of efflux pump overexpression on the resistance rates to antibiotics (detailed statistical analysis provided in the supplementary file). The overexpression of the MexEF-OprN pumps significantly induced the resistance of bacteria to cephalosporin (cefepime and ceftazidime), aminosides (gentamicin), and floroquinolone (ciprofloxacin and levofloxacin). Conversly, the floroquinolone resistance was found to be associated with MexCD-OprJ overexpression. Specifically, overexpression of MexEF-OprN was present in the majority of cases with strains resistant to tested antibiotic classes, such as carbapenem (58.8%–61.5%), cephalosporin (75.0%–78.3%), aminoglycoside (63.2%–80.0%), and fluoroquinolone (88.9%–90.0%). This result may suggest the substantial role of MexEF-OprN efflux pump in the antibiotic resistance of P. aeruginosa strains isolated from clinical samples in this study.
The number of strains that resisted against Piperaclin/tazobactam and Colistin was low and not reliable for analysis by Chi-square test, thus these data were removed from the Table.
1) The total number of strains that overexpressed certain types of efflux pump, without considering antibiotic resistance properties.
2) p-values were calculated by Pearson’s Chi-squared test with Yates’ continuity correction or Fisher’s exact test for count data.
4. DISCUSSION
Antibiotic resistance is a global health emergency and is particularly prevalent in developing countries, including Vietnam. Antibiotic resistance not only leads to treatment failures, increased complications, and mortality but also increases treatment costs, imposing a burden on patients and society. Therefore, alongside monitoring the rational use of antibiotics, monitoring antibiotic resistance in hospitals through antibiotic susceptibility testing is crucial for controlling antibiotic resistance. P. aeruginosa is among the pathogens closely monitored for antibiotic resistance rates in clinical settings. In this study, P. aeruginosa exhibited high resistance levels to most tested antibiotics, ranging from 38.33% to 65.00%. Particularly, the carbapenem antibiotic class showed high resistance rates, with meropenem at 56.67% and imipenem at 65.00%. Piperacillin-tazobactam and colistin were two antibiotics with consistently high sensitivity rates (>80%). The high resistance rates of P. aeruginosa strains to antibiotics align with findings from recent domestic studies [28-33] (Table 9).
| Study design | Findings | Author |
|---|---|---|
| P. aeruginosa from Hospital-acquired pneumonia at Da Nang C Hospital in 2022 (cross-sectional study) | P. aeruginosa accounted for 32.58% cases of HAP, and showed resistance against all of tested antibiotics: tobramycin (34.78%), gentamycin (34.62%), ciprofloxacin (50%), levofloxacin (57.69%), imipenem (48.15%), meropenem (44%), cefepime (44.44%) and ceftazidime (44%). Piperacillin/tazobactam and amikacin maintained their efficacy with the resistance rate as low as 10.35% and 15.38%, respectively. | Hoa et al. (2023) [28] |
| P. aeruginosa from exacerbaction of chronic obstructive pulmonary disease at Kien Giang Province General Hospital in 2021 (retrospective cross-sectional study) | P. aeruginosa was responsible for 17.39% of infection cases. These strains resisted against imipenem (28.57%), meropenem (21.43%), ciprofloxacin (29%), piperacillin/tazobactam (21.43%), amikacin (21.43%) | Linh et al. (2021) [29] |
| P. aeruginosa from Hospital-acquired infection at Nguyen Tri Phuong Hospital from 2019–2021 (retrospective cross-sectional study) | P. aeruginosa has shown complete resistance to co-trimexazole. Although the average sensitivity rate of these bacteria to commonly used antipseudomonal antibiotics such as beta-lactams and aminoglycosides remains above 50%, meropenem exhibits a lower sensitivity rate compared to antibiotics in the same group, such as imipenem (42.0% versus 54.3%). The quinolone group has sensitivity rates below 50%. | Ha et al. (2023) [30] |
| P. aeruginosa from lower respiratory tract infection at Cho Ray Hospital 2021 (retrospective cross-sectional study) | P. aeruginosa was isolated at a rate of 13.9%, exhibited the highest resistant to ticarcillin/clavilanic acid (78%) and carbapenem group (68%–70%), colistin resistant rate was 2%. | Phu et al. (2022) [31] |
| P. aeruginosa isolated at the 108 Military Central Hospital from 2020–2022 (cross-sectional study) | P. aeruginosa are highly resistant to fluoroquinolones (62.8%) and aminoglycosides (53.4%). Among the multidrug-resistant (MDR) strains, 54.3% were also resistant to carbapenem (CPR), with the highest proportion found in the department of infectious diseases (65.52%), ICU (64.39%) and the Internal respiratory department (45.69%). In the CPR strains, 11.2% of isolates were found to be resistant to colistin, while 26.6% and 33.3% of these strains remained susceptible to amikacin and piperacillin/tazobactam, respectively. | Trang et al.(2022) [32] |
| P. aeruginosa from Hospital-acquired infection at Binh Dan Hospital from 2018–2020 (cross-sectional study) | P. aeruginosa was isolated from 11.16% of patients and was found to resistant to more than 50% of most used antibiotics | Ngan & Phuong (2022) [33] |
The high resistance to multiple antibiotics of P. aeruginosa is attributed to the combination of various resistance mechanisms, such as reduced cell permeability to antibiotics, secretion of antibiotic-degrading enzymes, or target site mutations of antibiotics. Overexpression of efflux pumps is considered a crucial resistance mechanism, present in multidrug-resistant strains isolated from clinical settings, as documented in several studies (Table 10) [34-38]. The prevalence of antibiotic efflux pump overexpression in clinical strains of P. aeruginosa varies across studies, with the overexpression of MexAB-OprM and MexXY-OprM often being the most frequently observed [34-36]. However, Shigemura et al. (2015) reported higher expression levels of MexEF-OprN and MexXY-OprM compared to other pumps [37]. In our study, MexEF-OprN and MexCD-OprJ were the two efflux pump types with the highest overexpression rates, at 53.33% and 21.67%, respectively. This finding aligns with the results of Li (2000), suggesting an inverse relationship between the expression levels of MexEF-OprN or MexCD-OprJ efflux pump systems and the expression level of MexAB-OprM [39]. This feature indicates the overall complement of these MDR efflux systems, and that alterations in the level of one efflux system may affect compensatory changes in the levels of the others.
| Sample type | Efflux pump overexpression rates | Association with antibiotic susceptibility profile | Reference |
|---|---|---|---|
| P. aeruginosa from bloodstream infections | MexAB-OprM: 12.3% MexCD-OprJ: 2.2% MexEF-OprN: 4.2% MexXY-OprM: 13.2% |
Cefepime-resistant isolates: 25% overexpressed MexAB-OprM and 29% overexpressed MexXY-OprM. MexXY-OprM was the most prevalent efflux pump in tobramycin-resistant isolates (37%), while MexAB-OprM was the most common in meropenem-resistant isolates (33%). |
Gabriel et al. (2011) [34] |
| P. aeruginosa from bloodstream infections | MexAB-OprM: 50.8% MexXY-OprM: 27.1% |
MexAB-OprM overexpressing isolates showed high resistance to amikacin, gentamicin, and ciprofloxacin (86.7%), while MexXY-OprM overexpressing isolates showed high resistance to cefepime (80%). | Xavier et al. (2010) [35] |
| Ciprofloxacin-resistant P. aeruginosa | MexAB-OprM: 36.5% MexEF-OprN: 11.76% MexXY-OprM: 45.88% |
MexEF-OprN may be the main resistance mechanism of P. aeruginosa to fluoroquinolone antibiotics. | Llanes et al. (2011) [36] |
| P. aeruginosa from urinary tract infections | MexAB-OprM: 26.7% MexCD-OprJ: 11.4% MexEF-OprN: 41.9% MexXY-OprM: 38.1% |
MexCD-OprJ overexpression was associated with levofloxacin resistance. | Shigemura et al. (2015) [37] |
| Hospital-acquired P. aeruginosa infections | MexAB-OprM: 69% MexCD-OprJ: 28.7% MexEF-OprN: 43.4% MexXY-OprM: 74.6% |
Overexpression of MexAB-OprM and MexXY-OprM efflux pumps was highly correlated with bacterial resistance to the treatment antibiotics. | Zahedi bialvaei et al. (2021) [38] |
| Hospital-acquired P. aeruginosa infections | MexAB-OprM: 10.0% MexCD-OprJ: 21.67% MexEF-OprN: 53.33% MexXY-OprM: 5.0% |
The overexpression of MexEF-OprN was closely related to the resistance of baceria against cephalosporins, aminosides, and floroquinolones. The resistance of floroquinolone was also found to be related to MexCD-OprJ overexpression | This study |
The differences in the overexpression rates of efflux pumps reported across studies may arise from the accumulation of different mutation types under the pressure of antibiotics and/or the complexity of the resistance induced by horizontal gene transfer via mobile genetic elements. MexAB-OprM and MexXY-OprM are two types of efflux pumps naturally expressed at low levels in wild-type strains of P. aeruginosa. Mutations in regulatory genes such as mexR [40], nalC [41], or nalD [42] contribute to the overexpression of MexAB-OprM, while mexZ [43,44], parR, and parS [45,46] mutations affect MexEF-OprN expression. MexCD-OprJ is scarcely expressed in wild-type strains due to tight regulation by nfxB, and mutations in this regulatory gene lead to increased MexCD-OprJ expression [47,48]. Additionally, Sanz-García et al. found that prolonged exposure of P. aeruginosa strains to low concentrations of ciprofloxacin significantly increased the expression of MexCD and induced cross-resistance to other antibiotics [49]. Variants of the MexCD-OprJ encoding genes have also been identified in mobile genetic elements on chromosomes or plasmids present in P. aeruginosa and some other Gram-negative bacteria, indicating the complex relationship between MexCD-OprJ efflux pump overexpression and antibiotic resistance in clinical settings [50-52]. Similar to MexCD-OprJ, MexEF-OprN is hardly expressed in wild-type strains. Mutations in the mexT or mexS regulatory genes increase MexEF-OprN expression in resistant strains [16,53,54]. Therefore, the varied selection pressures resulting from different antibiotic usage frequencies in studied regions may be the main causes of inconsistent efflux pump overexpression rates among studies. Consequently, monitoring the rates of efflux pump overexpression should be considered as a crucial periodic survey, aiding in the surveillance of antibiotic resistance in clinical P. aeruginosa strains.
In this study, the increased expression of MexEF-OprN was closely associated with the antibiotic resistance profile of P. aeruginosa strains causing hospital infections, especially to cephalosporins, aminosides and floroquinolones. The resistance to floroquinolone was also found to be related to MexCD-OprJ overexpression. Similar findings were reported by Llanes et al. [36] and Shigemura et al. [37], where MexEF-OprN and MexCD-OprJ were closely linked to fluoroquinolone resistance. This explains why over 90% of fluoroquinolones – resistant strains in the study had MexEF-OprN and MexCD-OprJ. Furthermore, the augmented expression of MexEF-OprN is associated with decreased OprD expression, as mexT acts as an inducer for MexEF-OprN but an inhibitor for OprD. The reduced expression of the porin OprD is closely associated with carbapenem resistance, as it is the primary transport channel for this class of antibiotics [16,55,56]. Therefore, although not the direct target of MexEF-OprN, carbapenems may become less active in strains with increased MexEF-OprN expression. Thus, the high prevalence of overexpression of MexEF-OprN (58.8%–61.5%) in strains resistant to carbapenems observed in our study can be reasonably explained.
Although the phenotypic method used in this study cannot fully differentiate bacterial resistance mechanisms, it provides a preliminary assessment of whether efflux pump mechanisms are the primary contributors to bacterial resistance through the reversal of resistance in the presence of the inhibitor PaβN. Our results showed that 10 out of 25 (40%) clinical isolates of P. aeruginosa that overexpressed MexEF-OprN had their resistance reversed in the presence of the inhibitor PaβN (50 mg/L). This suggests that MexEF-OprN may be the main contributor to the antibiotic resistance of these strains. Therefore, the significance of increased expression of efflux pumps in bacteria and the need for research in screening EPIs should be acknowledged at a higher level.
5. CONCLUSION
This study effectively utilized a phenotypic method to assess the prevalence of overexpression of the four major RND efflux pumps in P. aeruginosa. Our findings also highlights the clinical significance of MexEF-OprN and MexCD-OprJ in contributing to the resistance profile of the studied strains.








